Prediction and Analysis of Crabs’ Age and Sex Base on Physical Features#
Author: Kaijie Zhang
Course Project, UC Irvine, Math 10, S23
Introduction#
Based on daily experiences, distinguishing between male and female crabs is crucial. Gender dictates a crab’s role in the market; for instance, female crabs are often used to produce crab roe due to their higher roe content. However, identifying a crab’s sex requires experience, especially for non-industrial fishermen. It is said that seasoned fishermen can estimate a crab’s gender by evaluating its weight and size. I intend to start with the age of the crabs: using a regressor to predict the crabs’ age based on available data. Subsequently, I will attempt to develop a machine learning model that aids fishermen in accurately determining the sex of crabs and assess its efficacy.
from PIL import Image
Image.open("dataset-cover.jpeg")

Section 1: Data Cleaning and Feature Engineering#
import pandas as pd
import numpy as np
import altair as alt
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.tree import plot_tree
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import train_test_split
import seaborn as sns
from itertools import product
from pandas.api.types import is_numeric_dtype
from sklearn.neighbors import KNeighborsRegressor, KNeighborsClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import mean_squared_error, mean_absolute_error
from sklearn.cluster import KMeans
df = pd.read_csv("Crab_Features.csv")
Have a short preview of the dataset:
df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 200000 entries, 0 to 199999
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 id 200000 non-null int64
1 Sex 200000 non-null object
2 Length 200000 non-null float64
3 Diameter 200000 non-null float64
4 Height 200000 non-null float64
5 Weight 200000 non-null float64
6 Shucked Weight 200000 non-null float64
7 Viscera Weight 200000 non-null float64
8 Shell Weight 200000 non-null float64
9 Age 200000 non-null float64
dtypes: float64(8), int64(1), object(1)
memory usage: 15.3+ MB
Since the original dataset is so large that Deepnote could not process all of them. I pick the first 5000 data sample to be our investigated dataset.
df=df[:5000]
df.dropna(inplace=True)
df.shape
(5000, 10)
Original dataset gives multiple plain but key variables. We can create our own features base on them:
df['Volume'] = df['Length'] * df['Diameter'] * df['Height']
# Crab BMI
# Water Loss during experiment
df["water_loss"]=df["Weight"]-df["Shucked Weight"]-df['Viscera Weight']-df['Shell Weight']
df["water_loss"]=np.where( df["water_loss"]<0,
min(df["Shucked Weight"].min(),
df["Viscera Weight"].min(), df["Shell Weight"].min()),
df["water_loss"])
# Crab density approx
df['Density'] = df['Weight']/(df['Volume'])
# Normalize weights to represent them as a ratio of the total weight
df['Shucked Weight Ratio'] = df['Shucked Weight'] / df['Weight']
df['Shell Weight Ratio'] = df['Shell Weight'] / df['Weight']
df['Viscera Weight Ratio'] = df['Viscera Weight'] / df['Weight']
df['water_loss Ratio'] = df['water_loss'] / df['Weight']
df['Sex_num'] = df['Sex'].replace({'I': 0, 'M': 1, 'F': 2})
/shared-libs/python3.7/py-core/lib/python3.7/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
"""Entry point for launching an IPython kernel.
/shared-libs/python3.7/py-core/lib/python3.7/site-packages/ipykernel_launcher.py:5: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
"""
/shared-libs/python3.7/py-core/lib/python3.7/site-packages/ipykernel_launcher.py:9: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
if __name__ == "__main__":
/shared-libs/python3.7/py-core/lib/python3.7/site-packages/ipykernel_launcher.py:11: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
# This is added back by InteractiveShellApp.init_path()
/shared-libs/python3.7/py-core/lib/python3.7/site-packages/ipykernel_launcher.py:14: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
/shared-libs/python3.7/py-core/lib/python3.7/site-packages/ipykernel_launcher.py:15: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
from ipykernel import kernelapp as app
/shared-libs/python3.7/py-core/lib/python3.7/site-packages/ipykernel_launcher.py:16: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
/shared-libs/python3.7/py-core/lib/python3.7/site-packages/ipykernel_launcher.py:17: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
app.launch_new_instance()
/shared-libs/python3.7/py-core/lib/python3.7/site-packages/ipykernel_launcher.py:19: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
Volume & Density: more familiar variables to replace height, length, and diameter. It will be a little confused if we use three variables to represent the “size” of the crabs.
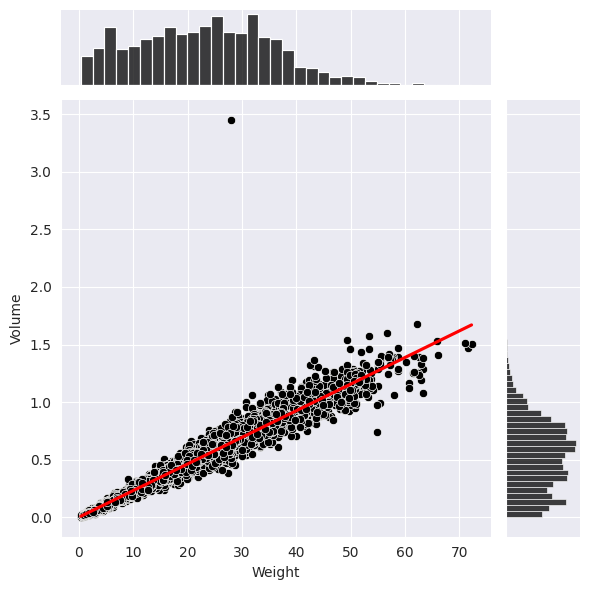
This is the relation between volume and weight (in the below plot by Seaborn), which is obviousily linear according to its regression line. Such combanation of various variables could help us review the whoe dataset more effectively. (We can see there is an outlier at about (29,3.45))
with sns.axes_style('darkgrid'):
# Create jointplot without regression line
joint = sns.jointplot(data=df, x='Weight', y='Volume', color='black')
# Add the regression line separately
joint.ax_joint.clear() # Clear the original points
sns.scatterplot(data=df, x='Weight', y='Volume', ax=joint.ax_joint, color='black')
sns.regplot(data=df, x='Weight', y='Volume', ax=joint.ax_joint, color='red', scatter=False)
Shucked/Viscera/Shell Weight ratio: the ratio of sections of each individual might implies more than just posting the weight of them respectively. [It will be used later to explain the water_loss]
water_loss: We observe that the addition of Shucked/Viscera/Shell Weight does not equal to the original weight of the individual. One significant reason is that the water contained in alive crabs will outflow when researchers are decomposing the crabs. The significant part of lost weight equals to the weight of water loss. (As you can see, water loss is actually visible in total weight [by ratio])
# Melt the DataFrame to long format
df_melt = df.copy().melt(id_vars='Age', value_vars=['Shucked Weight Ratio', 'Shell Weight Ratio', 'Viscera Weight Ratio', 'water_loss Ratio'], var_name='Weight Type', value_name='Weight Ratio')
# Due to the deepnote has a limitation of 5000 rows for dataframe, we'll aggregate data by taking mean of the ratios for each Age.
df_agg = df_melt.groupby(['Age', 'Weight Type']).mean().reset_index()
# Create the stacked bar chart
chart = alt.Chart(df_agg).mark_bar().encode(
x='Age:O',
y=alt.Y('Weight Ratio:Q', stack='normalize'),
color='Weight Type:N',
tooltip = ['Weight Ratio:Q']
).properties(
title="Ratio of Crabs' Weight")
chart
id is not related to our prediction, so I drop it to avoid any mistakes.
df.drop("id", axis=1)
| Sex | Length | Diameter | Height | Weight | Shucked Weight | Viscera Weight | Shell Weight | Age | Shucked Weight ratio | Viscera Weight ratio | Shell Weight ratio | Volume | water_loss | Density | Sex_num | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | M | 1.5750 | 1.2250 | 0.3750 | 31.226974 | 12.303683 | 6.321938 | 9.638830 | 10.0 | 0.394008 | 0.202451 | 0.308670 | 0.723516 | 2.962523 | 43.160055 | 1 |
| 1 | I | 1.2375 | 1.0000 | 0.3750 | 21.885814 | 7.654365 | 3.798833 | 7.654365 | 19.0 | 0.349741 | 0.173575 | 0.349741 | 0.464063 | 2.778251 | 47.161350 | 0 |
| 2 | F | 1.4500 | 1.1625 | 0.4125 | 28.250277 | 11.127179 | 7.016501 | 7.257472 | 11.0 | 0.393879 | 0.248369 | 0.256899 | 0.695320 | 2.849125 | 40.629155 | 2 |
| 3 | I | 1.3500 | 1.0250 | 0.3750 | 21.588144 | 9.738053 | 4.110678 | 6.378637 | 9.0 | 0.451083 | 0.190414 | 0.295469 | 0.518906 | 1.360776 | 41.603169 | 0 |
| 4 | I | 1.1375 | 0.8750 | 0.2875 | 14.968536 | 5.953395 | 2.962523 | 3.713785 | 8.0 | 0.397727 | 0.197917 | 0.248106 | 0.286152 | 2.338834 | 52.309675 | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 4995 | M | 1.6750 | 1.2750 | 0.5000 | 37.407165 | 14.855138 | 7.002326 | 11.906790 | 14.0 | 0.397120 | 0.187192 | 0.318302 | 1.067813 | 3.642911 | 35.031586 | 1 |
| 4996 | M | 1.6000 | 1.2000 | 0.4375 | 31.397071 | 14.047177 | 7.016501 | 8.930093 | 10.0 | 0.447404 | 0.223476 | 0.284424 | 0.840000 | 1.403300 | 37.377466 | 1 |
| 4997 | M | 1.6000 | 1.2750 | 0.4375 | 36.273185 | 15.521351 | 9.270286 | 10.404267 | 10.0 | 0.427902 | 0.255569 | 0.286831 | 0.892500 | 1.077281 | 40.642224 | 1 |
| 4998 | F | 1.3625 | 1.0375 | 0.3375 | 21.559795 | 9.128539 | 4.479221 | 7.087375 | 10.0 | 0.423406 | 0.207758 | 0.328731 | 0.477088 | 0.864660 | 45.190404 | 2 |
| 4999 | F | 1.4500 | 1.1625 | 0.3625 | 27.144646 | 10.517665 | 6.251065 | 7.512618 | 13.0 | 0.387467 | 0.230287 | 0.276762 | 0.611039 | 2.863299 | 44.423750 | 2 |
5000 rows × 16 columns
df.columns #Preview all current variables in the dataset:
Index(['id', 'Sex', 'Length', 'Diameter', 'Height', 'Weight', 'Shucked Weight',
'Viscera Weight', 'Shell Weight', 'Age', 'Shucked Weight ratio',
'Viscera Weight ratio', 'Shell Weight ratio', 'Volume', 'water_loss',
'Density', 'Sex_num'],
dtype='object')
Since Sex is the only String variable in the dataset, I prefer to separate that into three Boolean variables: Male, Female, and Indeterminate.
df_num = pd.get_dummies(df, columns=['Sex'])
Let see whether all columns in df_num are currently numeric:
len([c for c in df_num.columns if is_numeric_dtype(df_num[c])]) == df_num.shape[1]
True
df_num.sample(3)
| id | Length | Diameter | Height | Weight | Shucked Weight | Viscera Weight | Shell Weight | Age | Shucked Weight ratio | Viscera Weight ratio | Shell Weight ratio | Volume | water_loss | Density | Sex_num | Sex_F | Sex_I | Sex_M | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4697 | 4697 | 1.1250 | 0.8750 | 0.2750 | 12.998246 | 5.499803 | 2.806601 | 3.685435 | 9.0 | 0.423119 | 0.215921 | 0.283533 | 0.270703 | 1.006407 | 48.016608 | 0 | 0 | 1 | 0 |
| 4217 | 4217 | 1.4375 | 1.1875 | 0.4500 | 29.852024 | 10.347568 | 6.265239 | 11.765042 | 16.0 | 0.346629 | 0.209877 | 0.394112 | 0.768164 | 1.474174 | 38.861521 | 1 | 0 | 0 | 1 |
| 2978 | 2978 | 1.2875 | 0.9875 | 0.2875 | 16.768729 | 7.342521 | 3.614561 | 4.677668 | 10.0 | 0.437870 | 0.215554 | 0.278952 | 0.365529 | 1.133980 | 45.875199 | 2 | 1 | 0 | 0 |
Section 2: Regressor - Age Prediction#
A basic view of distribution of popuation in different age. We could see the majority of population locate in range from 7 yrs to 11 yrs.
alt.Chart(df).mark_bar().encode(
x = alt.X("Age", bin=alt.Bin(maxbins=100)),
y="count()"
).properties(
title="Distribution of Age")
First,before predicting the sex, I want to use regressor to predict the age according to crab’s weight by connecting PolynomialFeatures and LinearRegression model by Pipeline so that we can see different regression lines with different degrees.
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import PolynomialFeatures
from sklearn.linear_model import LinearRegression
The first stage uses the PolynomialFeatures class from sklearn.preprocessing to transform the original variable ‘Weight’ into polynomial features of a specified degree d. The argument include_bias=False ensures that a column of ones (the bias or intercept) is not added to the polynomial features. The second stage uses the LinearRegression class from sklearn.linear_model to perform linear regression on the transformed data.
def poly_fit(df,d,color):
df_sub = df.copy()
X = df_sub[["Weight"]]
y = df_sub["Age"]
pipe = Pipeline(
[
("poly", PolynomialFeatures(degree=d, include_bias=False)),
("reg", LinearRegression())
]
)
pipe.fit(X,y)
df_sub[f"pred{d}"] = pipe.predict(df_sub[["Weight"]])
chart = alt.Chart(df_sub).mark_line(clip=True, color=color).encode(
x=alt.X("Weight", scale=alt.Scale(domain=(0,80))),
y=alt.Y(f"pred{d}", scale=alt.Scale(domain=(0,28))),
)
return chart
chartl = alt.Chart(df).mark_circle().encode(
x="Weight",
y="Age"
)
I choose three representative degrees number: 1,2,8. Overly high degrees will cause overfitting to each data point, which makes the plot looks messy.
tstate_colors = [(1, 'red'), (2, 'blue'), (8, 'green')]
chart_list = [poly_fit(df,d,color) for d, color in state_colors]
We can observe that regression line in different degrees implies how many curves it might have: a staright line for 1 degrees, 2 curves for 2 degrees, and multiple curves for 8 degrees. But all of them could explain the positive relationship between weight and age (it could be slightly negative when crabs are old).
chartl+chart_list[0]+chart_list[1]+chart_list[2]
