Week 9 Wednesday#
Announcements#
Midterm 2 is Friday. Get a notecard if you need one.
Jinghao will go over the sample midterm (posted on the Week 9 page) in discussion on Thursday. Sample solutions will be posted after discussion section.
Worksheet 17 is posted. It is meant to get you started on the course project.
Worksheet 18 is also posted. I recommend finishing this worksheet before Midterm 2, as it is mostly review material. Worksheet 18 is the last worksheet for Math 10.
I have office hours starting at 12pm in here before the midterm on Friday.
The specific new functions we introduce today will not be on Midterm 2, but the concepts are relevant.
import pandas as pd
import altair as alt
Temperature data set#
Here is the dataset we finished with on Friday. We have succeeded in plotting the data, but we haven’t done any Machine Learning with the data yet.
We would really like to use a line like color="city", so that temperatures in different cities get different colors. We can’t do that with the current version of the DataFrame, because the city names are given in column names (as opposed to being given in their own column).
df_pre = pd.read_csv("temperature_f.csv", parse_dates=["datetime"])
df_pre.sample(4, random_state=0)
| datetime | Vancouver | Portland | San Francisco | Seattle | Los Angeles | San Diego | Las Vegas | Phoenix | Albuquerque | ... | Philadelphia | New York | Montreal | Boston | Beersheba | Tel Aviv District | Eilat | Haifa | Nahariyya | Jerusalem | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 37037 | 2016-12-22 18:00:00 | 38.7 | 31.7 | 52.9 | 34.9 | 55.3 | 60.9 | 45.6 | 60.8 | 34.1 | ... | 46.8 | 43.6 | 32.0 | 36.4 | 44.3 | 58.4 | 57.5 | 55.4 | 55.4 | 58.7 |
| 23197 | 2015-05-26 02:00:00 | 61.6 | 60.6 | 56.2 | 60.3 | 65.7 | 64.0 | 75.3 | 78.4 | 60.3 | ... | 75.0 | 73.2 | 62.5 | 66.9 | 58.2 | 59.7 | 71.4 | 69.9 | 69.9 | 65.1 |
| 33027 | 2016-07-08 16:00:00 | 59.9 | 62.0 | 59.3 | 59.9 | 70.9 | 70.5 | 90.6 | 95.2 | 79.6 | ... | 87.7 | 83.8 | 69.4 | 66.5 | 87.3 | 87.4 | 104.4 | 97.0 | 79.0 | 87.4 |
| 22542 | 2015-04-28 19:00:00 | 50.6 | 55.1 | 57.0 | 56.8 | 79.2 | 82.3 | 74.1 | 78.5 | 51.3 | ... | 63.6 | 63.6 | 58.7 | 50.8 | 75.0 | 70.7 | 74.1 | 70.7 | 66.9 | 70.6 |
4 rows × 37 columns
Originally the city names appear as column headers. For both Altair and scikit-learn, we want a column containing all these city names.
df_melted = df_pre.melt(
id_vars="datetime",
var_name="city",
value_name="temperature"
)
By using the method melt, we have gotten a much longer version of the DataFrame, that now has just three columns. The “city” column contains the old column headers (repeated many times each city), and the “temperature” column contains the old cell contents.
df_melted.sample(4, random_state=0)
| datetime | city | temperature | |
|---|---|---|---|
| 1478577 | 2016-03-25 22:00:00 | Eilat | 78.0 |
| 1299334 | 2016-06-07 11:00:00 | Montreal | 61.1 |
| 1072194 | 2016-05-01 19:00:00 | Miami | 84.5 |
| 863753 | 2013-03-15 18:00:00 | Atlanta | 48.5 |
The resulting DataFrame is much longer.
Here we check using an f-string. We use triple quotation marks so that we are allowed to include a line break in the string.
print(f"""The original DataFrame had shape {df_pre.shape}.
The new DataFrame has shape {df_melted.shape}.""")
The original DataFrame had shape (45252, 37).
The new DataFrame has shape (1629072, 3).
Here is what we did on Friday, although I’m naming the final DataFrame df3 instead of df, because I want to make more changes before calling the result df.
Keep only the rows corresponding to the cities in the following
citieslist.Drop the rows with missing values.
Sort the DataFrame by date.
Name the resulting DataFrame
df3.
cities = ["San Diego", "San Francisco", "Detroit"]
We keep only the rows corresponding to those three cities using Boolean indexing and the isin method. We then drop the rows with missing values. We then sort the values by date. (Probably the sorting would not be correct if the dates were strings instead of datetime objects.)
df1 = df_melted[df_melted["city"].isin(cities)]
df2 = df1.dropna(axis=0)
df3 = df2.sort_values("datetime")
Here is how the resulting DataFrame df3 looks. Notice how the first three rows are all from the same row and at 1pm. Then in the last row it moves to 2pm. (It does seem like the DataFrame is sorted by date, and we are only seeing the three listed cities, unlike earlier when we saw many different cities.)
df3.head(4)
| datetime | city | temperature | |
|---|---|---|---|
| 90504 | 2012-10-01 13:00:00 | San Francisco | 61.4 |
| 226260 | 2012-10-01 13:00:00 | San Diego | 65.1 |
| 905040 | 2012-10-01 13:00:00 | Detroit | 51.6 |
| 905041 | 2012-10-01 14:00:00 | Detroit | 51.7 |
Plotting the data#
Now that we have “melted” the DataFrame, it is easy to plot using Altair.
Our goal is to predict these values using scikit-learn. This will be our first time using a decision tree for regression (as opposed to classification).
c_true = alt.Chart(df3[6400:7000]).mark_line().encode(
x="datetime",
y="temperature",
color="city",
tooltip=["city", "temperature", "datetime"]
).properties(
width=600
)
c_true
Adding Boolean columns for the cities#
Because the “city” column contains strings, we can’t use it as an input when fitting a decision tree. We will convert it to multiple Boolean indicator columns.
Here are two options:
Use
pd.get_dummies.Use scikit-learn’s
OneHotEncoderclass.
I couldn’t immediately get OneHotEncoder to work with Pipeline (according to ChatGPT, it seems to require yet another tool called ColumnTransformer), so I’m going to use the pandas option.
Make a new DataFrame
df4containing Boolean indicator columns for the “city” values using the pandas functionget_dummies.
I couldn’t remember the name of the keyword argument I wanted to use, so I called the following help function. This reminded me that columns was the keyword argument I wanted to use.
help(pd.get_dummies)
Help on function get_dummies in module pandas.core.reshape.reshape:
get_dummies(data, prefix=None, prefix_sep='_', dummy_na=False, columns=None, sparse=False, drop_first=False, dtype=None) -> 'DataFrame'
Convert categorical variable into dummy/indicator variables.
Parameters
----------
data : array-like, Series, or DataFrame
Data of which to get dummy indicators.
prefix : str, list of str, or dict of str, default None
String to append DataFrame column names.
Pass a list with length equal to the number of columns
when calling get_dummies on a DataFrame. Alternatively, `prefix`
can be a dictionary mapping column names to prefixes.
prefix_sep : str, default '_'
If appending prefix, separator/delimiter to use. Or pass a
list or dictionary as with `prefix`.
dummy_na : bool, default False
Add a column to indicate NaNs, if False NaNs are ignored.
columns : list-like, default None
Column names in the DataFrame to be encoded.
If `columns` is None then all the columns with
`object` or `category` dtype will be converted.
sparse : bool, default False
Whether the dummy-encoded columns should be backed by
a :class:`SparseArray` (True) or a regular NumPy array (False).
drop_first : bool, default False
Whether to get k-1 dummies out of k categorical levels by removing the
first level.
dtype : dtype, default np.uint8
Data type for new columns. Only a single dtype is allowed.
Returns
-------
DataFrame
Dummy-coded data.
See Also
--------
Series.str.get_dummies : Convert Series to dummy codes.
Examples
--------
>>> s = pd.Series(list('abca'))
>>> pd.get_dummies(s)
a b c
0 1 0 0
1 0 1 0
2 0 0 1
3 1 0 0
>>> s1 = ['a', 'b', np.nan]
>>> pd.get_dummies(s1)
a b
0 1 0
1 0 1
2 0 0
>>> pd.get_dummies(s1, dummy_na=True)
a b NaN
0 1 0 0
1 0 1 0
2 0 0 1
>>> df = pd.DataFrame({'A': ['a', 'b', 'a'], 'B': ['b', 'a', 'c'],
... 'C': [1, 2, 3]})
>>> pd.get_dummies(df, prefix=['col1', 'col2'])
C col1_a col1_b col2_a col2_b col2_c
0 1 1 0 0 1 0
1 2 0 1 1 0 0
2 3 1 0 0 0 1
>>> pd.get_dummies(pd.Series(list('abcaa')))
a b c
0 1 0 0
1 0 1 0
2 0 0 1
3 1 0 0
4 1 0 0
>>> pd.get_dummies(pd.Series(list('abcaa')), drop_first=True)
b c
0 0 0
1 1 0
2 0 1
3 0 0
4 0 0
>>> pd.get_dummies(pd.Series(list('abc')), dtype=float)
a b c
0 1.0 0.0 0.0
1 0.0 1.0 0.0
2 0.0 0.0 1.0
Here is a reminder of how the top of the data looks.
df3.head(4)
| datetime | city | temperature | |
|---|---|---|---|
| 90504 | 2012-10-01 13:00:00 | San Francisco | 61.4 |
| 226260 | 2012-10-01 13:00:00 | San Diego | 65.1 |
| 905040 | 2012-10-01 13:00:00 | Detroit | 51.6 |
| 905041 | 2012-10-01 14:00:00 | Detroit | 51.7 |
Here is the result of calling the get_dummies function for the “city” column. This is a common procedure in Machine Learning. We have a column with data we want to use as an input (in this case, we want to use the city), but we aren’t able to because the data is not numeric. Here we add a column for each city, indicating if the row corresponded to that particular city. For example, notice that our DataFrame begins “not Detroit”, “not Detroit”, “Detroit”, “Detroit”. This corresponds to the “city_Detroit” column beginning [0, 0, 1, 1]. I think of this as being equivalent to [False, False, True, True].
The main point is that we can use these three new columns as inputs to our Machine Learning algorithm.
pd.get_dummies(df3, columns=["city"])
| datetime | temperature | city_Detroit | city_San Diego | city_San Francisco | |
|---|---|---|---|---|---|
| 90504 | 2012-10-01 13:00:00 | 61.4 | 0 | 0 | 1 |
| 226260 | 2012-10-01 13:00:00 | 65.1 | 0 | 1 | 0 |
| 905040 | 2012-10-01 13:00:00 | 51.6 | 1 | 0 | 0 |
| 905041 | 2012-10-01 14:00:00 | 51.7 | 1 | 0 | 0 |
| 90505 | 2012-10-01 14:00:00 | 61.4 | 0 | 0 | 1 |
| ... | ... | ... | ... | ... | ... |
| 271509 | 2017-11-29 22:00:00 | 67.0 | 0 | 1 | 0 |
| 950290 | 2017-11-29 23:00:00 | 40.8 | 1 | 0 | 0 |
| 271510 | 2017-11-29 23:00:00 | 67.0 | 0 | 1 | 0 |
| 271511 | 2017-11-30 00:00:00 | 64.8 | 0 | 1 | 0 |
| 950291 | 2017-11-30 00:00:00 | 38.2 | 1 | 0 | 0 |
134964 rows × 5 columns
We are doing the same thing in the next cell, the only difference being that we save the result with the variable name df4.
df4 = pd.get_dummies(df3, columns=["city"])
Make a list
city_colscontaining the three new column names we just added.
Here is how we can make that list using list comprehension.
city_cols = [c for c in df4.columns if c.startswith("city_")]
city_cols
['city_Detroit', 'city_San Diego', 'city_San Francisco']
The following alternative approach doesn’t seem as robust, because we need to know there are three columns, and we need to know they’re at the end of the DataFrame. On the other hand, with the following approach, we don’t need to know that the column names start with "city_".
df4.columns[-3:]
Index(['city_Detroit', 'city_San Diego', 'city_San Francisco'], dtype='object')
Add the “city” column back in (so we can use it with plotting) using
pd.concatwith an appropriateaxiskeyword argument. Name the resulting DataFramedf_ext(for “extended”).
Unfortunately the following error is a little difficult to interpret. It is implying that df3[["city"]] is being treated like an axis argument.
pd.concat(df4, df3[["city"]], axis=1)
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
/tmp/ipykernel_106/1418832173.py in <module>
----> 1 pd.concat(df4, df3[["city"]], axis=1)
TypeError: concat() got multiple values for argument 'axis'
The DataFrames we want to merge together are supposed to be grouped together, for example in a list or a tuple. Here we put them into a tuple.
Notice also that we use the keyword argument axis=1, because we are changing the column labels and keeping the row labels the same. (We are including a new column label, the label “city”.)
df_ext = pd.concat((df4, df3[["city"]]), axis=1)
df_ext
| datetime | temperature | city_Detroit | city_San Diego | city_San Francisco | city | |
|---|---|---|---|---|---|---|
| 90504 | 2012-10-01 13:00:00 | 61.4 | 0 | 0 | 1 | San Francisco |
| 226260 | 2012-10-01 13:00:00 | 65.1 | 0 | 1 | 0 | San Diego |
| 905040 | 2012-10-01 13:00:00 | 51.6 | 1 | 0 | 0 | Detroit |
| 905041 | 2012-10-01 14:00:00 | 51.7 | 1 | 0 | 0 | Detroit |
| 90505 | 2012-10-01 14:00:00 | 61.4 | 0 | 0 | 1 | San Francisco |
| ... | ... | ... | ... | ... | ... | ... |
| 271509 | 2017-11-29 22:00:00 | 67.0 | 0 | 1 | 0 | San Diego |
| 950290 | 2017-11-29 23:00:00 | 40.8 | 1 | 0 | 0 | Detroit |
| 271510 | 2017-11-29 23:00:00 | 67.0 | 0 | 1 | 0 | San Diego |
| 271511 | 2017-11-30 00:00:00 | 64.8 | 0 | 1 | 0 | San Diego |
| 950291 | 2017-11-30 00:00:00 | 38.2 | 1 | 0 | 0 | Detroit |
134964 rows × 6 columns
Predicting temperature using DecisionTreeRegressor#
We want to use the “datetime” column and the “city” column. We need to convert the “city” column to (multiple) Boolean columns before we can use it as an input.
Make a list
colscontaining the columns incity_colsand invar_cols.
It’s easy to concatenate two lists: just use +. This new list cols will be our input features.
cols = city_cols + var_cols
cols
['city_Detroit',
'city_San Diego',
'city_San Francisco',
'year',
'day',
'month',
'hour',
'day_of_week']
Fit a
DecisionTreeRegressorobject with a maximum of16leaf nodes to the data in these columns (notice that they’re all numeric). Use “temperature” as our target.
This is our first time using a DecisionTreeRegressor, but it works very similarly to DecisionTreeClassifier. The main difference is that the target values in the regression case should be numbers.
from sklearn.tree import DecisionTreeRegressor
Here we instantiate the regressor, specifying the maximum number of leaf nodes.
reg = DecisionTreeRegressor(max_leaf_nodes=16)
Here we fit the regressor, using “temperature” as our target.
reg.fit(df[cols], df["temperature"])
DecisionTreeRegressor(max_leaf_nodes=16)
Add a “pred” column to
dfcontaining the predicted temperatures.
df["pred"] = reg.predict(df[cols])
Plot the data using the following.
alt.Chart(df[6400:7000]).mark_line().encode(
x="datetime",
y="pred",
color="city",
tooltip=["city", "temperature", "pred", "datetime", "hour"]
).properties(
width=600
)
This is just one small portion of the data, corresponding to about one week. Notice how we only see San Francisco and Detroit. That is because the predictions for San Francisco and San Diego are the same.
Also, because we used a maximum of 16 leaf nodes, there are a maximum of 16 values that can be predicted. That is why we see so many flat line segments in this chart.
alt.Chart(df[6400:7000]).mark_line().encode(
x="datetime",
y="pred",
color="city",
tooltip=["city", "temperature", "pred", "datetime", "hour"]
).properties(
width=600
)
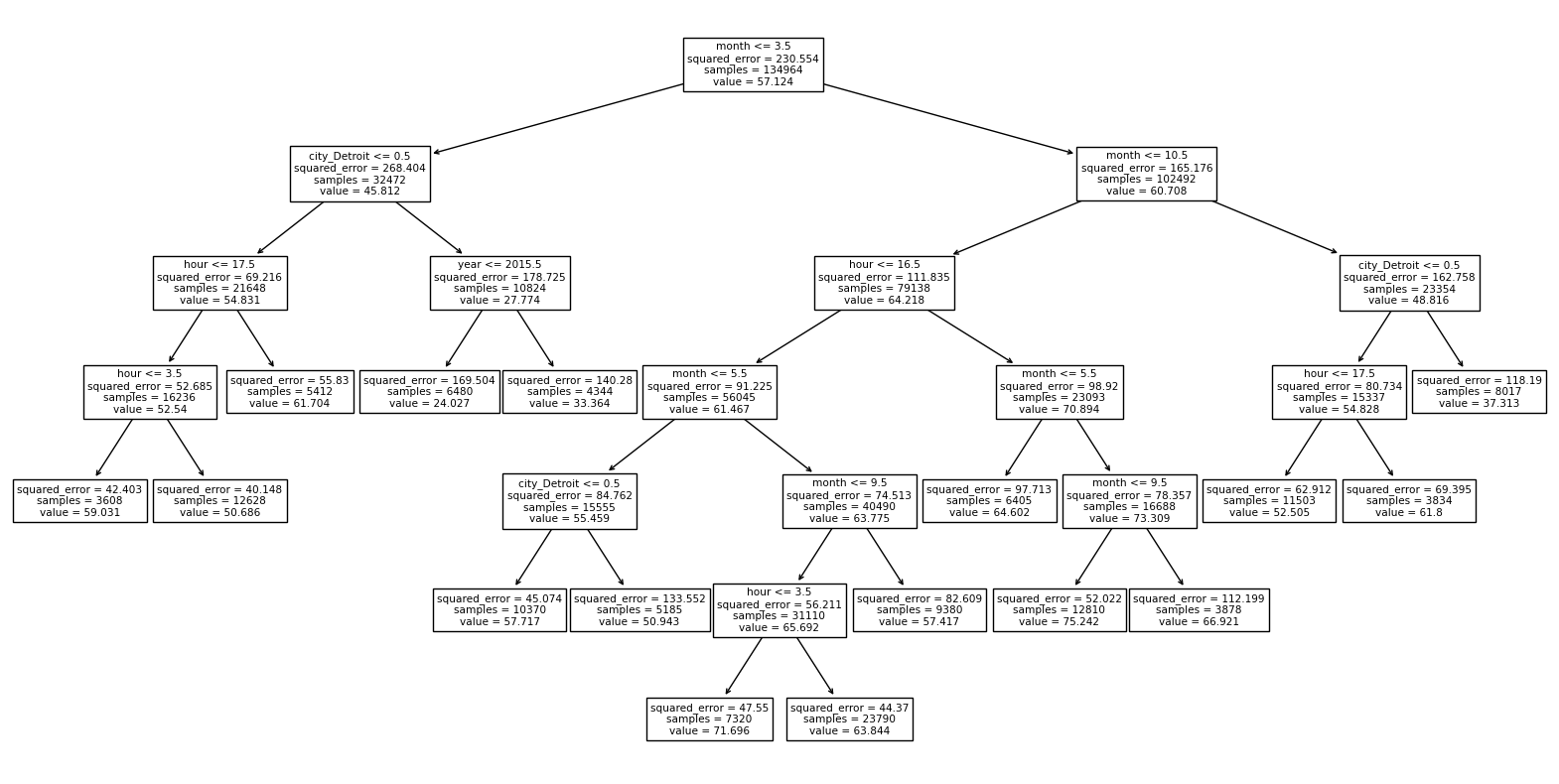
Can you see where the predicted values come from, using the following diagram?
(We will need to change clf to something else.)
import matplotlib.pyplot as plt
from sklearn.tree import plot_tree
fig = plt.figure(figsize=(20,10))
_ = plot_tree(clf,
feature_names=clf.feature_names_in_,
filled=False)
Using the Altair tooltip, I focused on a point for San Francisco on January 1st at 7am. Following the diagram, we wind up near the lower-left portion of the diagram, with value 50.686. This is indeed the predicted temperature for the point we were looking at. For the particular point I was looking at, the true temperature was 42.3.
import matplotlib.pyplot as plt
from sklearn.tree import plot_tree
fig = plt.figure(figsize=(20,10))
_ = plot_tree(reg,
feature_names=reg.feature_names_in_,
filled=False)
How does the Altair chart change if you use a max depth of
14instead of a max leaf nodes of16? (Don’t try to make the tree diagram in this case.)
This might sound similar, but it is extremely different.
reg = DecisionTreeRegressor(max_depth=14)
reg.fit(df[cols], df["temperature"])
DecisionTreeRegressor(max_depth=14)
df["pred"] = reg.predict(df[cols])
Here is the chart. Recall how above the same predictions were made for San Diego and San Francisco (at least in the part of the chart we are plotting). Here there are many different values plotted.
c_14 = alt.Chart(df[6400:7000]).mark_line().encode(
x="datetime",
y="pred",
color="city",
tooltip=["city", "temperature", "pred", "datetime", "hour"]
).properties(
width=600
)
c_14
How do those values compare to the true values? Look how similar they are! Is that a good sign? No, it is almost surely a sign of overfitting. Using a depth of 14 in this case is providing too much flexibility to the model.
c_true + c_14
How many leaf nodes are there in this case? Use the method
get_n_leaves.
reg.get_n_leaves()
12722
The theoretical maximum number of leaf nodes for a depth of 14 is \(2^{14}\).
2**14
16384
Do you think this new decision tree would do well at predicting unseen data?
No, I would expect poor performance on new data, because it is overfitting the input data.
I don’t think using
train_test_splitmakes much sense on this data, do you see why?
Imagine we separate the data into a training set and a test set. Maybe the training set includes the data from 7am (for a particular city and day) and the test set includes the data from 8am (for that same city and day). In real life, the temperatures for 7am and 8am will be very similar, so we will probably get a very accurate prediction for the 8am value, but I do not think that is meaningful. (Another way to think about it, is if we want to predict the temperature at some day in the future, we will not know what the temperature is one hour earlier.)
In short, if 7am is in the training set, it’s like we already knew the 8am value.
I believe a better way with this sort of data (this is called time series data): remove a whole month or a whole year, and see how we do on the predictions for that missing month or year. That performance would be much more indicative of how we might hope to do on new predictions in the future.
We didn’t get to the following.
Extra time?#
I doubt we will get here, but if we do, this will give a sense of how decision trees make their predictions. (Not how they make their cuts.)
Make a new decision tree regressor that allows only 4 leaf nodes.
Draw the corresponding diagram using
plot_tree.
Can you recover the given value for one of the leaf nodes using Boolean indexing?
