Week 10 Wednesday#
Announcements#
The Course Project is due a week from today.
Make sure you’re completing the project in the Project Template file in the Course Project folder on Deepnote. (Don’t complete your project on the Worksheet 17 template.)
My goal today is to show how I could use Bing chat to help me learn a new topic (how to use
OneHotEncoderas part of a scikit-learn pipeline).
Cautions#
Bing chat and ChatGPT are amazing tools, but you should use them with caution.
Bing chat always sounds confident, but is often wrong.
Even when Bing chat is correct, it is often using outdated methods. (Several times I have learned how to code something from Bing chat, and later found a much simpler way elsewhere, such as on StackOverflow.)
Last I checked, Bing chat is nearly useless for solving mathematical problems (unless it is a classic problem where the solution can be found verbatim on the internet).
Setup#
I want to perform linear regression on the cars dataset from Seaborn, using “mpg” as the target. I want to include information from the “origin” column among the input features. Because the “origin” column contains strings, we will need to convert it into (multiple) numeric columns. We did this with city names using pd.get_dummies. Today I want to use scikit-learn’s OneHotEncoder.
import seaborn as sns
import pandas as pd
df = sns.load_dataset("mpg")
df.head(4)
| mpg | cylinders | displacement | horsepower | weight | acceleration | model_year | origin | name | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 18.0 | 8 | 307.0 | 130.0 | 3504 | 12.0 | 70 | usa | chevrolet chevelle malibu |
| 1 | 15.0 | 8 | 350.0 | 165.0 | 3693 | 11.5 | 70 | usa | buick skylark 320 |
| 2 | 18.0 | 8 | 318.0 | 150.0 | 3436 | 11.0 | 70 | usa | plymouth satellite |
| 3 | 16.0 | 8 | 304.0 | 150.0 | 3433 | 12.0 | 70 | usa | amc rebel sst |
df["origin"].value_counts()
usa 249
japan 79
europe 70
Name: origin, dtype: int64
With help from Bing chat#
In the following screenshots, you can see my questions (on the right side) and Bing’s responses on the left side. (I made a mistake and asked about “mpg” when I meant “origin”. It doesn’t really make sense to use one-hot encoding with the “mpg” column, because there are so many different numbers in that column. One-hot encoding makes the most sense when there are a limited number of values.)

from sklearn.preprocessing import OneHotEncoder
enc = OneHotEncoder()
This OneHotEncoder works pretty similarly to PolynomialFeatures which we have used earlier in this class. One difference is that we didn’t need to pass keyword arguments when instantiating the object, unlike for example PolynomialFeatures(degree=3, include_bias=False).
Maybe a more intimidating difference is that the output isn’t a traditional NumPy array. The reason behind this strange “sparse matrix” output is that “most” of the entries in the result will be 0, especially if there are many columns, so you can imagine possibly saving storage space by not actually storing the entire matrix at the same time.
enc.fit_transform(df[["origin"]])
<398x3 sparse matrix of type '<class 'numpy.float64'>'
with 398 stored elements in Compressed Sparse Row format>
But anyway, we can get a “traditional” NumPy array, by calling the toarray method.
arr = enc.fit_transform(df[["origin"]]).toarray()
arr
array([[0., 0., 1.],
[0., 0., 1.],
[0., 0., 1.],
...,
[0., 0., 1.],
[0., 0., 1.],
[0., 0., 1.]])
Let’s get a sense of what these numbers mean. Here are 5 random origin values.
df.sample(5, random_state=2)["origin"]
94 usa
32 usa
279 japan
178 europe
354 europe
Name: origin, dtype: object
Here are the same random 5 rows from the output of our enc.fit_transform call. (We got these rows by using the index attribute of the DataFrame we saw above. This only works because the index labels are the same as the row numbers.)
Notice how we have two “usa” values, then one “japan” value, then two “europe” values. We can tell from the following output, that the right-most column corresponds to “usa”, the middle column corresponds to “japan”, and the left-hand column corresponds to “europe”.
arr[df.sample(5, random_state=2).index]
array([[0., 0., 1.],
[0., 0., 1.],
[0., 1., 0.],
[1., 0., 0.],
[1., 0., 0.]])
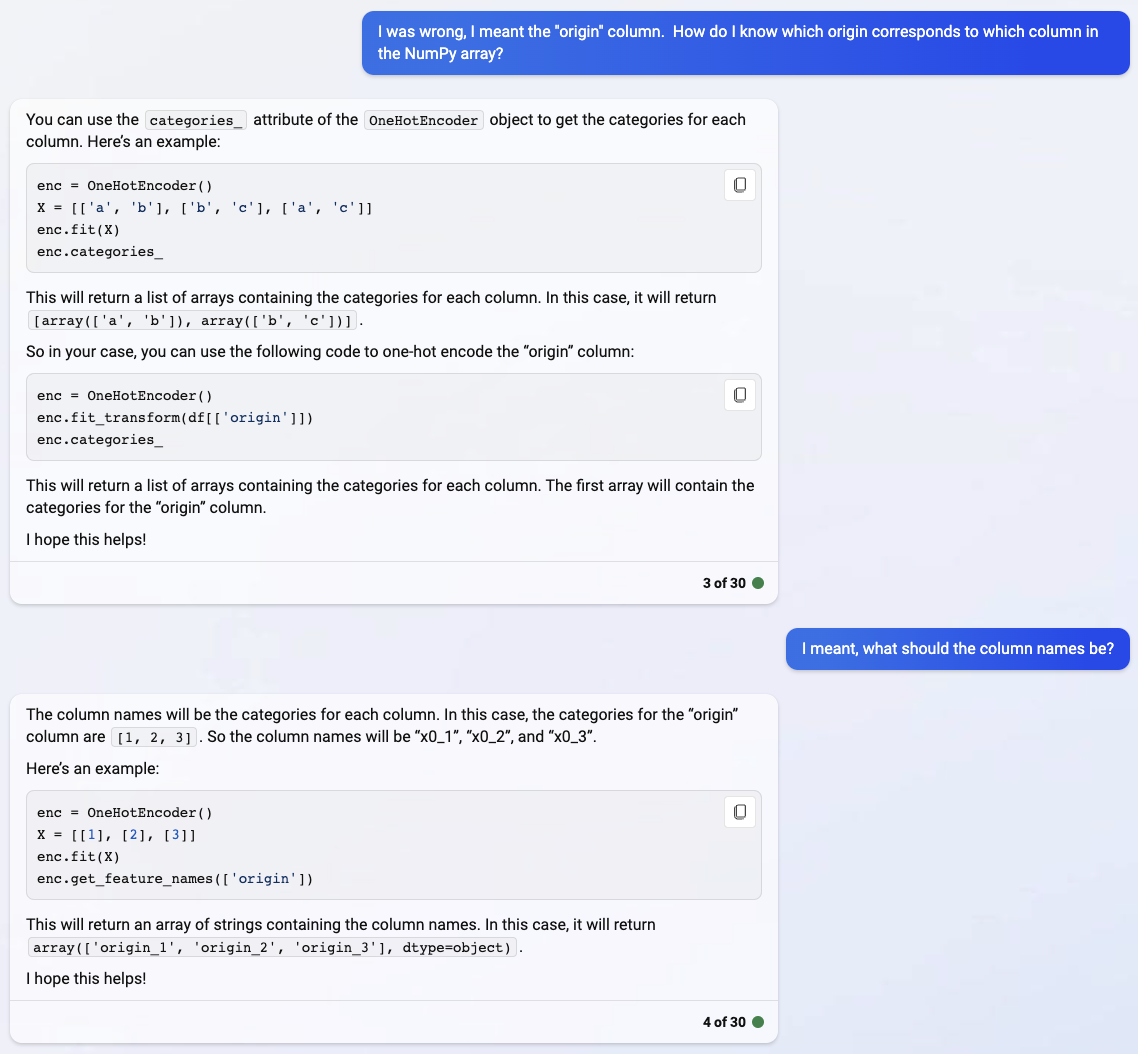
The answers Bing chat gives are often somewhat out-of-date. Here is an example. On Deepnote, I got a FutureWarning. Here on my personal computer, I’m getting an actual error. We are supposed to use a different method. The Deepnote version instructed me to use get_feature_names_out. (The below error does not give instructions on what to do.)
enc.get_feature_names(["origin"])
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[12], line 1
----> 1 enc.get_feature_names(["origin"])
AttributeError: 'OneHotEncoder' object has no attribute 'get_feature_names'
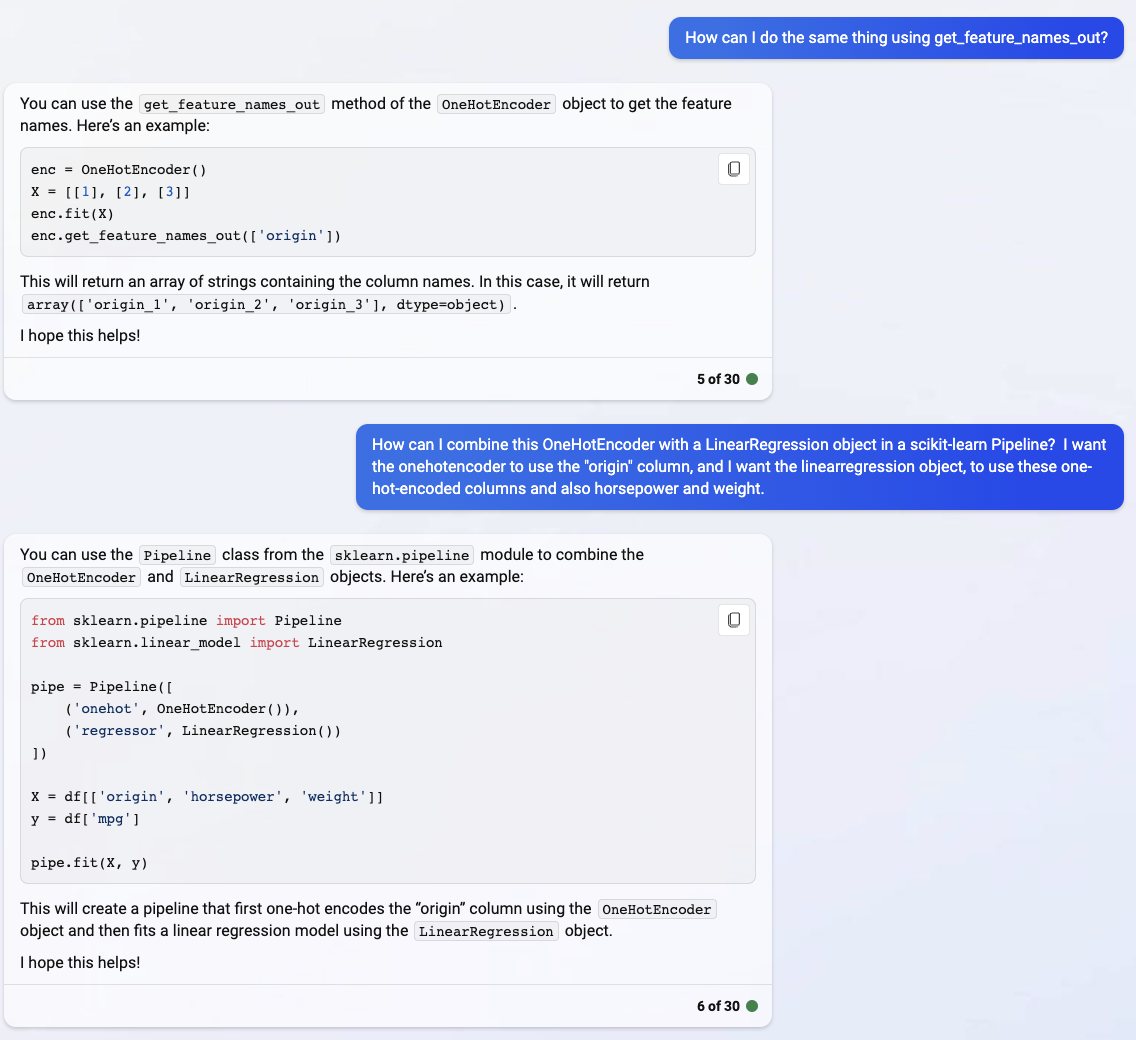
Notice how these values are in the same order as we saw above.
enc.get_feature_names_out(["origin"])
array(['origin_europe', 'origin_japan', 'origin_usa'], dtype=object)

The main reason I didn’t introduce OneHotEncoder earlier (and instead used pd.get_dummies), was because getting OneHotEncoder into a Pipeline is not as straight-forward as getting a PolynomialFeatures into a Pipeline.
from sklearn.pipeline import Pipeline
from sklearn.linear_model import LinearRegression
pipe = Pipeline([
('onehot', OneHotEncoder()),
('regressor', LinearRegression())
])
X = df[['origin', 'horsepower', 'weight']]
y = df['mpg']
pipe.fit(X, y)
Pipeline(steps=[('onehot', OneHotEncoder()), ('regressor', LinearRegression())])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('onehot', OneHotEncoder()), ('regressor', LinearRegression())])OneHotEncoder()
LinearRegression()
Here we can see that the first suggestion above does not work as wanted. Notice how many “horsepower” and “weight” columns have been added. We only wanted to do this one-hot encoding with the “origin” column, not with the numerical columns.
pipe['onehot'].get_feature_names_out()
array(['origin_europe', 'origin_japan', 'origin_usa', 'horsepower_46.0',
'horsepower_48.0', 'horsepower_49.0', 'horsepower_52.0',
'horsepower_53.0', 'horsepower_54.0', 'horsepower_58.0',
'horsepower_60.0', 'horsepower_61.0', 'horsepower_62.0',
'horsepower_63.0', 'horsepower_64.0', 'horsepower_65.0',
'horsepower_66.0', 'horsepower_67.0', 'horsepower_68.0',
'horsepower_69.0', 'horsepower_70.0', 'horsepower_71.0',
'horsepower_72.0', 'horsepower_74.0', 'horsepower_75.0',
'horsepower_76.0', 'horsepower_77.0', 'horsepower_78.0',
'horsepower_79.0', 'horsepower_80.0', 'horsepower_81.0',
'horsepower_82.0', 'horsepower_83.0', 'horsepower_84.0',
'horsepower_85.0', 'horsepower_86.0', 'horsepower_87.0',
'horsepower_88.0', 'horsepower_89.0', 'horsepower_90.0',
'horsepower_91.0', 'horsepower_92.0', 'horsepower_93.0',
'horsepower_94.0', 'horsepower_95.0', 'horsepower_96.0',
'horsepower_97.0', 'horsepower_98.0', 'horsepower_100.0',
'horsepower_102.0', 'horsepower_103.0', 'horsepower_105.0',
'horsepower_107.0', 'horsepower_108.0', 'horsepower_110.0',
'horsepower_112.0', 'horsepower_113.0', 'horsepower_115.0',
'horsepower_116.0', 'horsepower_120.0', 'horsepower_122.0',
'horsepower_125.0', 'horsepower_129.0', 'horsepower_130.0',
'horsepower_132.0', 'horsepower_133.0', 'horsepower_135.0',
'horsepower_137.0', 'horsepower_138.0', 'horsepower_139.0',
'horsepower_140.0', 'horsepower_142.0', 'horsepower_145.0',
'horsepower_148.0', 'horsepower_149.0', 'horsepower_150.0',
'horsepower_152.0', 'horsepower_153.0', 'horsepower_155.0',
'horsepower_158.0', 'horsepower_160.0', 'horsepower_165.0',
'horsepower_167.0', 'horsepower_170.0', 'horsepower_175.0',
'horsepower_180.0', 'horsepower_190.0', 'horsepower_193.0',
'horsepower_198.0', 'horsepower_200.0', 'horsepower_208.0',
'horsepower_210.0', 'horsepower_215.0', 'horsepower_220.0',
'horsepower_225.0', 'horsepower_230.0', 'horsepower_nan',
'weight_1613', 'weight_1649', 'weight_1755', 'weight_1760',
'weight_1773', 'weight_1795', 'weight_1800', 'weight_1825',
'weight_1834', 'weight_1835', 'weight_1836', 'weight_1845',
'weight_1850', 'weight_1867', 'weight_1875', 'weight_1915',
'weight_1925', 'weight_1937', 'weight_1940', 'weight_1945',
'weight_1950', 'weight_1955', 'weight_1963', 'weight_1965',
'weight_1968', 'weight_1970', 'weight_1975', 'weight_1980',
'weight_1985', 'weight_1990', 'weight_1995', 'weight_2000',
'weight_2003', 'weight_2019', 'weight_2020', 'weight_2025',
'weight_2035', 'weight_2045', 'weight_2046', 'weight_2050',
'weight_2051', 'weight_2065', 'weight_2070', 'weight_2074',
'weight_2075', 'weight_2085', 'weight_2100', 'weight_2108',
'weight_2110', 'weight_2120', 'weight_2123', 'weight_2124',
'weight_2125', 'weight_2126', 'weight_2130', 'weight_2135',
'weight_2144', 'weight_2145', 'weight_2150', 'weight_2155',
'weight_2158', 'weight_2160', 'weight_2164', 'weight_2171',
'weight_2188', 'weight_2189', 'weight_2190', 'weight_2200',
'weight_2202', 'weight_2205', 'weight_2210', 'weight_2215',
'weight_2219', 'weight_2220', 'weight_2223', 'weight_2226',
'weight_2228', 'weight_2230', 'weight_2234', 'weight_2245',
'weight_2246', 'weight_2254', 'weight_2255', 'weight_2264',
'weight_2265', 'weight_2278', 'weight_2279', 'weight_2288',
'weight_2290', 'weight_2295', 'weight_2300', 'weight_2310',
'weight_2320', 'weight_2330', 'weight_2335', 'weight_2350',
'weight_2370', 'weight_2372', 'weight_2375', 'weight_2379',
'weight_2380', 'weight_2385', 'weight_2391', 'weight_2395',
'weight_2401', 'weight_2405', 'weight_2408', 'weight_2420',
'weight_2430', 'weight_2434', 'weight_2451', 'weight_2464',
'weight_2472', 'weight_2489', 'weight_2490', 'weight_2500',
'weight_2506', 'weight_2511', 'weight_2515', 'weight_2525',
'weight_2542', 'weight_2545', 'weight_2556', 'weight_2560',
'weight_2565', 'weight_2572', 'weight_2575', 'weight_2582',
'weight_2585', 'weight_2587', 'weight_2592', 'weight_2595',
'weight_2600', 'weight_2605', 'weight_2615', 'weight_2620',
'weight_2625', 'weight_2634', 'weight_2635', 'weight_2639',
'weight_2640', 'weight_2648', 'weight_2660', 'weight_2665',
'weight_2670', 'weight_2671', 'weight_2672', 'weight_2678',
'weight_2694', 'weight_2700', 'weight_2702', 'weight_2711',
'weight_2720', 'weight_2725', 'weight_2735', 'weight_2740',
'weight_2745', 'weight_2755', 'weight_2774', 'weight_2789',
'weight_2790', 'weight_2795', 'weight_2800', 'weight_2807',
'weight_2815', 'weight_2830', 'weight_2833', 'weight_2835',
'weight_2855', 'weight_2865', 'weight_2868', 'weight_2870',
'weight_2875', 'weight_2890', 'weight_2900', 'weight_2901',
'weight_2904', 'weight_2905', 'weight_2910', 'weight_2914',
'weight_2930', 'weight_2933', 'weight_2945', 'weight_2950',
'weight_2957', 'weight_2962', 'weight_2965', 'weight_2979',
'weight_2984', 'weight_2990', 'weight_3003', 'weight_3012',
'weight_3015', 'weight_3021', 'weight_3035', 'weight_3039',
'weight_3060', 'weight_3070', 'weight_3085', 'weight_3086',
'weight_3102', 'weight_3121', 'weight_3139', 'weight_3140',
'weight_3150', 'weight_3155', 'weight_3158', 'weight_3160',
'weight_3169', 'weight_3190', 'weight_3193', 'weight_3205',
'weight_3210', 'weight_3211', 'weight_3221', 'weight_3230',
'weight_3233', 'weight_3245', 'weight_3250', 'weight_3264',
'weight_3265', 'weight_3270', 'weight_3278', 'weight_3282',
'weight_3288', 'weight_3302', 'weight_3329', 'weight_3336',
'weight_3353', 'weight_3360', 'weight_3365', 'weight_3380',
'weight_3381', 'weight_3399', 'weight_3410', 'weight_3415',
'weight_3420', 'weight_3425', 'weight_3430', 'weight_3432',
'weight_3433', 'weight_3436', 'weight_3439', 'weight_3445',
'weight_3449', 'weight_3459', 'weight_3465', 'weight_3504',
'weight_3520', 'weight_3525', 'weight_3530', 'weight_3535',
'weight_3563', 'weight_3570', 'weight_3574', 'weight_3605',
'weight_3609', 'weight_3613', 'weight_3620', 'weight_3630',
'weight_3632', 'weight_3645', 'weight_3651', 'weight_3664',
'weight_3672', 'weight_3693', 'weight_3725', 'weight_3730',
'weight_3735', 'weight_3755', 'weight_3761', 'weight_3777',
'weight_3781', 'weight_3785', 'weight_3820', 'weight_3821',
'weight_3830', 'weight_3840', 'weight_3850', 'weight_3870',
'weight_3880', 'weight_3892', 'weight_3897', 'weight_3900',
'weight_3907', 'weight_3940', 'weight_3955', 'weight_3962',
'weight_3988', 'weight_4042', 'weight_4054', 'weight_4055',
'weight_4060', 'weight_4077', 'weight_4080', 'weight_4082',
'weight_4096', 'weight_4098', 'weight_4100', 'weight_4129',
'weight_4135', 'weight_4140', 'weight_4141', 'weight_4154',
'weight_4165', 'weight_4190', 'weight_4209', 'weight_4215',
'weight_4220', 'weight_4237', 'weight_4257', 'weight_4274',
'weight_4278', 'weight_4294', 'weight_4295', 'weight_4312',
'weight_4325', 'weight_4335', 'weight_4341', 'weight_4354',
'weight_4360', 'weight_4363', 'weight_4376', 'weight_4380',
'weight_4382', 'weight_4385', 'weight_4422', 'weight_4425',
'weight_4440', 'weight_4456', 'weight_4457', 'weight_4464',
'weight_4498', 'weight_4499', 'weight_4502', 'weight_4615',
'weight_4633', 'weight_4638', 'weight_4654', 'weight_4657',
'weight_4668', 'weight_4699', 'weight_4732', 'weight_4735',
'weight_4746', 'weight_4906', 'weight_4951', 'weight_4952',
'weight_4955', 'weight_4997', 'weight_5140'], dtype=object)
The following error is not the fault of Bing, it’s because our data has missing values in it.
from sklearn.compose import ColumnTransformer
pipe = Pipeline([
('prep', ColumnTransformer([
('onehot', OneHotEncoder(), ['origin']),
('passthrough', 'passthrough', ['horsepower', 'weight'])
])),
('reg', LinearRegression())
])
X = df[['origin', 'horsepower', 'weight']]
y = df['mpg']
pipe.fit(X, y)
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[17], line 14
11 X = df[['origin', 'horsepower', 'weight']]
12 y = df['mpg']
---> 14 pipe.fit(X, y)
File ~/mambaforge/envs/math10s23/lib/python3.9/site-packages/sklearn/pipeline.py:405, in Pipeline.fit(self, X, y, **fit_params)
403 if self._final_estimator != "passthrough":
404 fit_params_last_step = fit_params_steps[self.steps[-1][0]]
--> 405 self._final_estimator.fit(Xt, y, **fit_params_last_step)
407 return self
File ~/mambaforge/envs/math10s23/lib/python3.9/site-packages/sklearn/linear_model/_base.py:648, in LinearRegression.fit(self, X, y, sample_weight)
644 n_jobs_ = self.n_jobs
646 accept_sparse = False if self.positive else ["csr", "csc", "coo"]
--> 648 X, y = self._validate_data(
649 X, y, accept_sparse=accept_sparse, y_numeric=True, multi_output=True
650 )
652 sample_weight = _check_sample_weight(
653 sample_weight, X, dtype=X.dtype, only_non_negative=True
654 )
656 X, y, X_offset, y_offset, X_scale = _preprocess_data(
657 X,
658 y,
(...)
661 sample_weight=sample_weight,
662 )
File ~/mambaforge/envs/math10s23/lib/python3.9/site-packages/sklearn/base.py:584, in BaseEstimator._validate_data(self, X, y, reset, validate_separately, **check_params)
582 y = check_array(y, input_name="y", **check_y_params)
583 else:
--> 584 X, y = check_X_y(X, y, **check_params)
585 out = X, y
587 if not no_val_X and check_params.get("ensure_2d", True):
File ~/mambaforge/envs/math10s23/lib/python3.9/site-packages/sklearn/utils/validation.py:1106, in check_X_y(X, y, accept_sparse, accept_large_sparse, dtype, order, copy, force_all_finite, ensure_2d, allow_nd, multi_output, ensure_min_samples, ensure_min_features, y_numeric, estimator)
1101 estimator_name = _check_estimator_name(estimator)
1102 raise ValueError(
1103 f"{estimator_name} requires y to be passed, but the target y is None"
1104 )
-> 1106 X = check_array(
1107 X,
1108 accept_sparse=accept_sparse,
1109 accept_large_sparse=accept_large_sparse,
1110 dtype=dtype,
1111 order=order,
1112 copy=copy,
1113 force_all_finite=force_all_finite,
1114 ensure_2d=ensure_2d,
1115 allow_nd=allow_nd,
1116 ensure_min_samples=ensure_min_samples,
1117 ensure_min_features=ensure_min_features,
1118 estimator=estimator,
1119 input_name="X",
1120 )
1122 y = _check_y(y, multi_output=multi_output, y_numeric=y_numeric, estimator=estimator)
1124 check_consistent_length(X, y)
File ~/mambaforge/envs/math10s23/lib/python3.9/site-packages/sklearn/utils/validation.py:921, in check_array(array, accept_sparse, accept_large_sparse, dtype, order, copy, force_all_finite, ensure_2d, allow_nd, ensure_min_samples, ensure_min_features, estimator, input_name)
915 raise ValueError(
916 "Found array with dim %d. %s expected <= 2."
917 % (array.ndim, estimator_name)
918 )
920 if force_all_finite:
--> 921 _assert_all_finite(
922 array,
923 input_name=input_name,
924 estimator_name=estimator_name,
925 allow_nan=force_all_finite == "allow-nan",
926 )
928 if ensure_min_samples > 0:
929 n_samples = _num_samples(array)
File ~/mambaforge/envs/math10s23/lib/python3.9/site-packages/sklearn/utils/validation.py:161, in _assert_all_finite(X, allow_nan, msg_dtype, estimator_name, input_name)
144 if estimator_name and input_name == "X" and has_nan_error:
145 # Improve the error message on how to handle missing values in
146 # scikit-learn.
147 msg_err += (
148 f"\n{estimator_name} does not accept missing values"
149 " encoded as NaN natively. For supervised learning, you might want"
(...)
159 "#estimators-that-handle-nan-values"
160 )
--> 161 raise ValueError(msg_err)
ValueError: Input X contains NaN.
LinearRegression does not accept missing values encoded as NaN natively. For supervised learning, you might want to consider sklearn.ensemble.HistGradientBoostingClassifier and Regressor which accept missing values encoded as NaNs natively. Alternatively, it is possible to preprocess the data, for instance by using an imputer transformer in a pipeline or drop samples with missing values. See https://scikit-learn.org/stable/modules/impute.html You can find a list of all estimators that handle NaN values at the following page: https://scikit-learn.org/stable/modules/impute.html#estimators-that-handle-nan-values
Here we drop the rows with missing values. I think this is safer than filling in the missing values.
df = df.dropna(axis=0)
Now we use the second suggestion shown in the screenshot above. I have not completely understood the meaning of these “passthrough” columns; at this point, I am following Bing’s instructions.
from sklearn.compose import ColumnTransformer
pipe = Pipeline([
('prep', ColumnTransformer([
('onehot', OneHotEncoder(), ['origin']),
('passthrough', 'passthrough', ['horsepower', 'weight'])
])),
('reg', LinearRegression())
])
X = df[['origin', 'horsepower', 'weight']]
y = df['mpg']
pipe.fit(X, y)
Pipeline(steps=[('prep',
ColumnTransformer(transformers=[('onehot', OneHotEncoder(),
['origin']),
('passthrough', 'passthrough',
['horsepower', 'weight'])])),
('reg', LinearRegression())])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('prep',
ColumnTransformer(transformers=[('onehot', OneHotEncoder(),
['origin']),
('passthrough', 'passthrough',
['horsepower', 'weight'])])),
('reg', LinearRegression())])ColumnTransformer(transformers=[('onehot', OneHotEncoder(), ['origin']),
('passthrough', 'passthrough',
['horsepower', 'weight'])])['origin']
OneHotEncoder()
['horsepower', 'weight']
passthrough
LinearRegression()
Remember that we can’t get the linear regression coefficients directly from the Pipeline object.
pipe.coef_
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[20], line 1
----> 1 pipe.coef_
AttributeError: 'Pipeline' object has no attribute 'coef_'
Instead, we have to access the LinearRegression step, which we do here using pipe["reg"].
pipe["reg"].coef_
array([-0.27333471, 1.50778661, -1.2344519 , -0.05354417, -0.00484275])
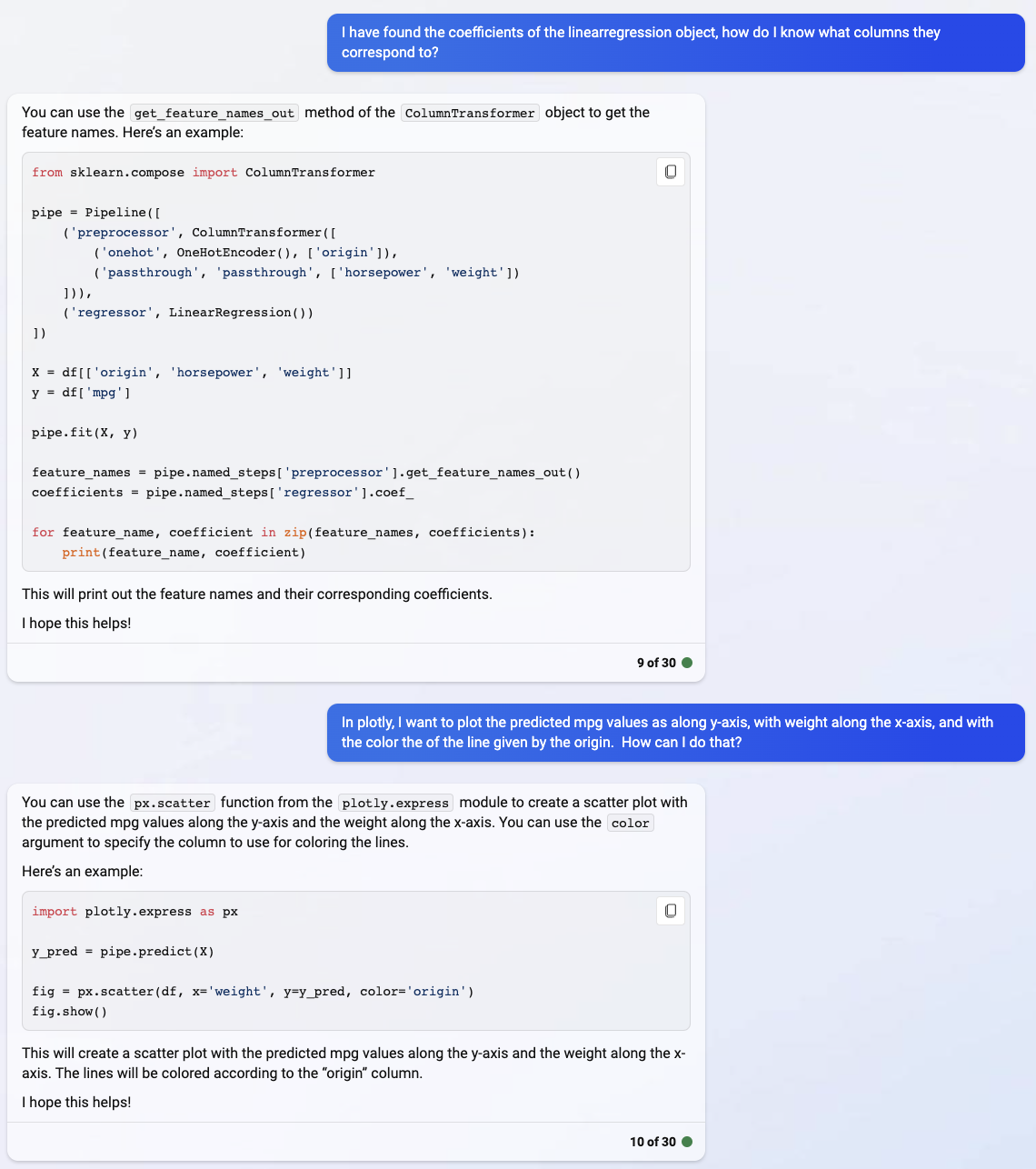
I’m not quite following the zip and print suggestion of Bing. But the main thing I wanted to learn was about getting the feature_names variable that Bing illustrates above, by calling the get_feature_names_out method on the preprocessor step.
pd.Series(
data=pipe["reg"].coef_,
index=pipe["prep"].get_feature_names_out()
)
onehot__origin_europe -0.273335
onehot__origin_japan 1.507787
onehot__origin_usa -1.234452
passthrough__horsepower -0.053544
passthrough__weight -0.004843
dtype: float64

Bing didn’t quite understand what I meant, and it gave me a scatterplot description.
Here we try plotting. It looks terrible, let’s try getting rid of the “horsepower” portion, since it’s not one of our axes here.
import plotly.express as px
y_pred = pipe.predict(X)
fig = px.line(df, x='weight', y=y_pred, color='origin')
fig.show()
I’m not sure what the error is below.
from sklearn.compose import ColumnTransformer
pipe = Pipeline([
('prep', ColumnTransformer([
('onehot', OneHotEncoder(), ['origin']),
('passthrough', ['weight'])
])),
('reg', LinearRegression())
])
X = df[['origin', 'weight']]
y = df['mpg']
pipe.fit(X, y)
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In [29], line 14
11 X = df[['origin', 'weight']]
12 y = df['mpg']
---> 14 pipe.fit(X, y)
File /shared-libs/python3.9/py/lib/python3.9/site-packages/sklearn/pipeline.py:378, in Pipeline.fit(self, X, y, **fit_params)
352 """Fit the model.
353
354 Fit all the transformers one after the other and transform the
(...)
375 Pipeline with fitted steps.
376 """
377 fit_params_steps = self._check_fit_params(**fit_params)
--> 378 Xt = self._fit(X, y, **fit_params_steps)
379 with _print_elapsed_time("Pipeline", self._log_message(len(self.steps) - 1)):
380 if self._final_estimator != "passthrough":
File /shared-libs/python3.9/py/lib/python3.9/site-packages/sklearn/pipeline.py:336, in Pipeline._fit(self, X, y, **fit_params_steps)
334 cloned_transformer = clone(transformer)
335 # Fit or load from cache the current transformer
--> 336 X, fitted_transformer = fit_transform_one_cached(
337 cloned_transformer,
338 X,
339 y,
340 None,
341 message_clsname="Pipeline",
342 message=self._log_message(step_idx),
343 **fit_params_steps[name],
344 )
345 # Replace the transformer of the step with the fitted
346 # transformer. This is necessary when loading the transformer
347 # from the cache.
348 self.steps[step_idx] = (name, fitted_transformer)
File /shared-libs/python3.9/py/lib/python3.9/site-packages/joblib/memory.py:349, in NotMemorizedFunc.__call__(self, *args, **kwargs)
348 def __call__(self, *args, **kwargs):
--> 349 return self.func(*args, **kwargs)
File /shared-libs/python3.9/py/lib/python3.9/site-packages/sklearn/pipeline.py:870, in _fit_transform_one(transformer, X, y, weight, message_clsname, message, **fit_params)
868 with _print_elapsed_time(message_clsname, message):
869 if hasattr(transformer, "fit_transform"):
--> 870 res = transformer.fit_transform(X, y, **fit_params)
871 else:
872 res = transformer.fit(X, y, **fit_params).transform(X)
File /shared-libs/python3.9/py/lib/python3.9/site-packages/sklearn/compose/_column_transformer.py:686, in ColumnTransformer.fit_transform(self, X, y)
684 # set n_features_in_ attribute
685 self._check_n_features(X, reset=True)
--> 686 self._validate_transformers()
687 self._validate_column_callables(X)
688 self._validate_remainder(X)
File /shared-libs/python3.9/py/lib/python3.9/site-packages/sklearn/compose/_column_transformer.py:346, in ColumnTransformer._validate_transformers(self)
343 if not self.transformers:
344 return
--> 346 names, transformers, _ = zip(*self.transformers)
348 # validate names
349 self._validate_names(names)
ValueError: not enough values to unpack (expected 3, got 2)
Honestly it still looks worse than I expected. The only difference between the origins is that they have different y-intercepts (or biases). In my head, I had expected them to have different slopes, but that is clearly wrong.
pipe = Pipeline([
('preprocessor', ColumnTransformer([
('onehot', OneHotEncoder(), ['origin']),
('passthrough', 'passthrough', ['weight'])
])),
('regressor', LinearRegression())
])
X = df[['origin', 'weight']]
y = df['mpg']
pipe.fit(X, y)
y_pred = pipe.predict(X)
fig = px.line(df, x='weight', y=y_pred, color='origin')
fig.show()
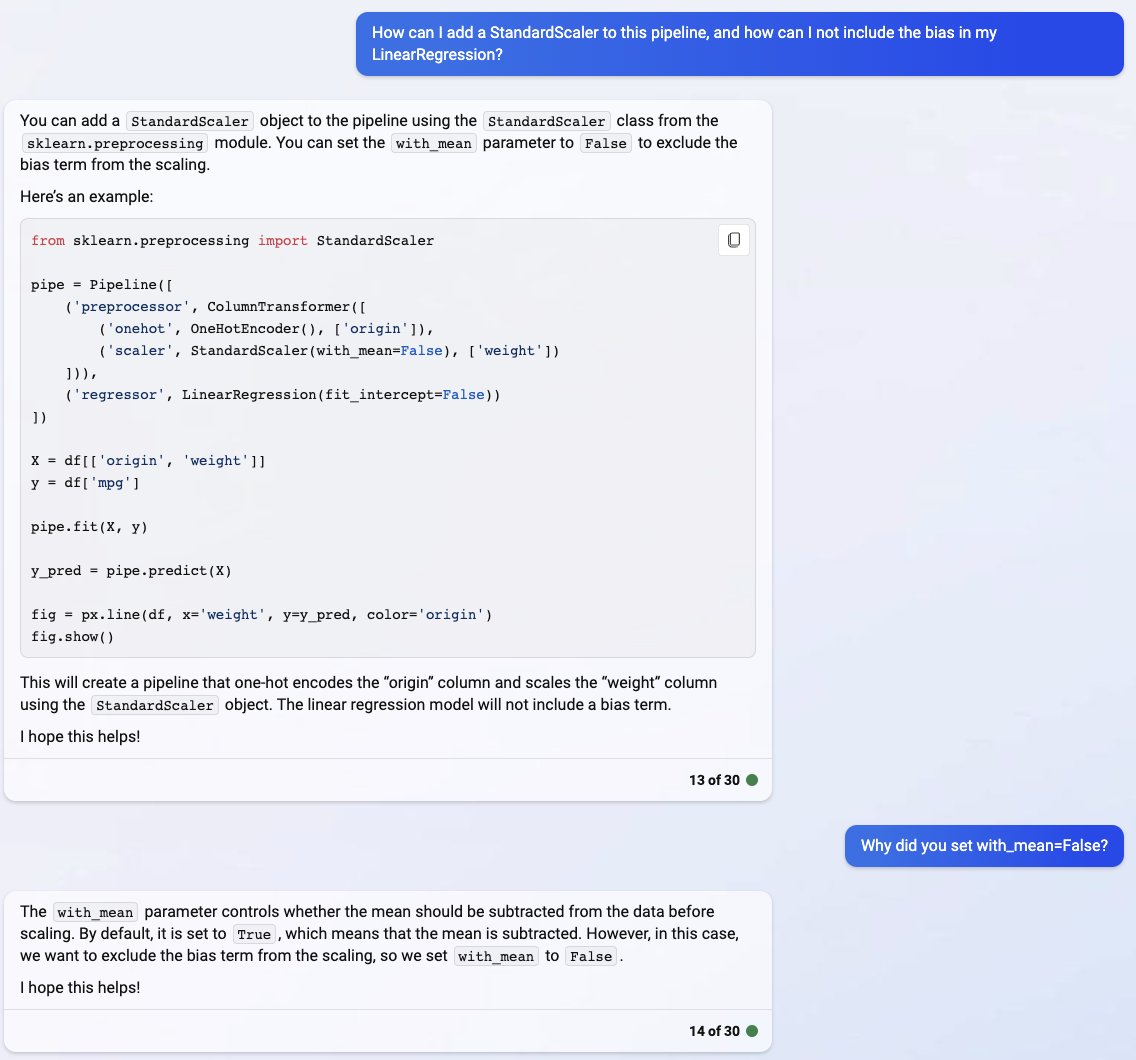
I don’t really think the with_mean part is correct, but I didn’t have time to investigate it during class.
from sklearn.preprocessing import StandardScaler
pipe = Pipeline([
('preprocessor', ColumnTransformer([
('onehot', OneHotEncoder(), ['origin']),
('scaler', StandardScaler(with_mean=False), ['weight'])
])),
('regressor', LinearRegression(fit_intercept=False))
])
X = df[['origin', 'weight']]
y = df['mpg']
pipe.fit(X, y)
y_pred = pipe.predict(X)
fig = px.line(df, x='weight', y=y_pred, color='origin')
fig.show()
Here are the corresponding coefficients.
pipe["regressor"].coef_
array([44.70314174, 46.05938605, 43.73223615, -5.9612044 ])
Notice how we don’t have an intercept_ any more (or more accurately, it is 0). That is because we instantiated the LinearRegression object using LinearRegression(fit_intercept=False).
pipe["regressor"].intercept_
0.0
