Zoo Animal Classification
Contents
Zoo Animal Classification¶
Author: Alisa Crowe
Course Project, UC Irvine, Math 10, S22
Introduction¶
In this project, I will be examining two datasets that hold data about 101 different species of zoo animals and various characteristics that they hold. I will be using Sci-Kit Learn’s K-Means Clustering, Logistic Regression, Decision Tree Classifier, and K-Nearest Neighbors to attempt to classify the animals while disucssion any results and/or challenges I come across. I will then analyze each of these methods to conclude which was the best in classifying.
Main portion of the project¶
Data Cleaning and Merging Dataframes¶
import pandas as pd
df_zoo = pd.read_csv("zoo.csv")
df_zoo
| animal_name | hair | feathers | eggs | milk | airborne | aquatic | predator | toothed | backbone | breathes | venomous | fins | legs | tail | domestic | catsize | class_type | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | aardvark | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 4 | 0 | 0 | 1 | 1 |
| 1 | antelope | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 4 | 1 | 0 | 1 | 1 |
| 2 | bass | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 4 |
| 3 | bear | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 4 | 0 | 0 | 1 | 1 |
| 4 | boar | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 4 | 1 | 0 | 1 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 96 | wallaby | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 1 | 1 |
| 97 | wasp | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 6 | 0 | 0 | 0 | 6 |
| 98 | wolf | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 4 | 1 | 0 | 1 | 1 |
| 99 | worm | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 7 |
| 100 | wren | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 0 | 2 |
101 rows × 18 columns
Here we introduce our first dataframe df_zoo. To ensure that there are no repeats of species, we can check value_counts() of the column ‘animal_name’.
df_zoo["animal_name"].value_counts()
frog 2
newt 1
aardvark 1
cavy 1
termite 1
..
crab 1
stingray 1
seahorse 1
bear 1
ladybird 1
Name: animal_name, Length: 100, dtype: int64
We can see that there are two rows for frogs in this dataframe, but only one of every other species.
df_zoo[df_zoo.loc[:,"animal_name"] == "frog"]
| animal_name | hair | feathers | eggs | milk | airborne | aquatic | predator | toothed | backbone | breathes | venomous | fins | legs | tail | domestic | catsize | class_type | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 25 | frog | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 4 | 0 | 0 | 0 | 4 |
| 26 | frog | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 4 | 0 | 0 | 0 | 4 |
Looking at the ‘venomous’ column, it is clear that these two rows are not duplicates; we can leave both of them in.
df_class = pd.read_csv("class.csv")
df_class
| Class_Number | Number_Of_Animal_Species_In_Class | Class_Type | Animal_Names | |
|---|---|---|---|---|
| 0 | 1 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... |
| 1 | 2 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... |
| 2 | 3 | 5 | Reptile | pitviper, seasnake, slowworm, tortoise, tuatara |
| 3 | 4 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... |
| 4 | 5 | 4 | Amphibian | frog, frog, newt, toad |
| 5 | 6 | 8 | Bug | flea, gnat, honeybee, housefly, ladybird, moth... |
| 6 | 7 | 10 | Invertebrate | clam, crab, crayfish, lobster, octopus, scorpi... |
Since I plan on merging these two dataframes together, I want to ensure that df_zoo["class_type"] matches up with the values in df_class["Number_Of_Animal_Species_In_Class"].
df_zoo["class_type"].value_counts()
0 41
1 20
3 13
6 10
5 8
2 5
4 4
Name: class_type, dtype: int64
To make things easier to read, I want to reset all the values in both df_zoo["class_type"] and df_class["Class_Number"] to be subtracted by one, so that it matches the index and starts at 0 instead of 1. I am doing this using a lambda function together with apply so that it applies to the entire column, and a lambda function is appropriate here because it is a very simple function.
df_zoo["class_type"] = df_zoo["class_type"].apply(lambda x: x-1)
df_class["Class_Number"] = df_class["Class_Number"].apply(lambda x: x-1)
Now to clean the data, I am using .isna().any().any() to see if there are any missing values in either dataframe. In this case, there are none.
# data cleaning
df_zoo.isna().any().any()
False
df_class.isna().any().any()
False
In order to make one dataframe, I am going to merge the two dataframes together with .merge. Since both dataframes have the same column with different names, they will be merged on this column. We can see that the resulting dataframe df has 101 columns; this is because I used how="left".
df = df_zoo.merge(df_class, how="left", left_on="class_type", right_on="Class_Number")
df
| animal_name | hair | feathers | eggs | milk | airborne | aquatic | predator | toothed | backbone | ... | fins | legs | tail | domestic | catsize | class_type | Class_Number | Number_Of_Animal_Species_In_Class | Class_Type | Animal_Names | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | aardvark | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 0 | 4 | 0 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... |
| 1 | antelope | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | ... | 0 | 4 | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... |
| 2 | bass | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | ... | 1 | 0 | 1 | 0 | 0 | 3 | 3 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... |
| 3 | bear | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 0 | 4 | 0 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... |
| 4 | boar | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 0 | 4 | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 96 | wallaby | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | ... | 0 | 2 | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... |
| 97 | wasp | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | ... | 0 | 6 | 0 | 0 | 0 | 5 | 5 | 8 | Bug | flea, gnat, honeybee, housefly, ladybird, moth... |
| 98 | wolf | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 0 | 4 | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... |
| 99 | worm | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 6 | 6 | 10 | Invertebrate | clam, crab, crayfish, lobster, octopus, scorpi... |
| 100 | wren | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | ... | 0 | 2 | 1 | 0 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... |
101 rows × 22 columns
Now that we are working with only one dataframe, we can check the dtypes to see which columns can be used for the various machine learning tequniques we are going to be using. Since all of the characteristic data (e.g. “hair”, “eggs”) is of type int64, these can be used.
df.dtypes
animal_name object
hair int64
feathers int64
eggs int64
milk int64
airborne int64
aquatic int64
predator int64
toothed int64
backbone int64
breathes int64
venomous int64
fins int64
legs int64
tail int64
domestic int64
catsize int64
class_type int64
Class_Number int64
Number_Of_Animal_Species_In_Class int64
Class_Type object
Animal_Names object
dtype: object
I will start by making a list of column names that are usable for the following machine learning techniques I will be using in this project. We want to use the columns that contain the characteristics of the animal which are the 1st through the 17th items in the list df.columns. Here I am using slicing on df.columns to obtain this list, naming it numcols.
df.columns
Index(['animal_name', 'hair', 'feathers', 'eggs', 'milk', 'airborne',
'aquatic', 'predator', 'toothed', 'backbone', 'breathes', 'venomous',
'fins', 'legs', 'tail', 'domestic', 'catsize', 'class_type',
'Class_Number', 'Number_Of_Animal_Species_In_Class', 'Class_Type',
'Animal_Names'],
dtype='object')
numcols = df.columns[1:17]
numcols
Index(['hair', 'feathers', 'eggs', 'milk', 'airborne', 'aquatic', 'predator',
'toothed', 'backbone', 'breathes', 'venomous', 'fins', 'legs', 'tail',
'domestic', 'catsize'],
dtype='object')
K-Means Clustering¶
I am using the standard process of importing, instantiating, fitting, and predicting for K-Means Clustering. Here I both fit and predict on df[numcols], making a new column df["pred"] for the predicted values. My goal for this section is to have the clusters match the class number for the animals as closely as possible.
# import
from sklearn.cluster import KMeans
# instatiate
kmeans = KMeans(n_clusters=7)
# fit
kmeans.fit(df[numcols])
KMeans(n_clusters=7)
# predict
df["pred"] = kmeans.predict(df[numcols])
df
| animal_name | hair | feathers | eggs | milk | airborne | aquatic | predator | toothed | backbone | ... | legs | tail | domestic | catsize | class_type | Class_Number | Number_Of_Animal_Species_In_Class | Class_Type | Animal_Names | pred | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | aardvark | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 4 | 0 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 |
| 1 | antelope | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | ... | 4 | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 |
| 2 | bass | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 3 | 3 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... | 2 |
| 3 | bear | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 4 | 0 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 |
| 4 | boar | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 4 | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 96 | wallaby | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | ... | 2 | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 4 |
| 97 | wasp | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | ... | 6 | 0 | 0 | 0 | 5 | 5 | 8 | Bug | flea, gnat, honeybee, housefly, ladybird, moth... | 3 |
| 98 | wolf | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 4 | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 |
| 99 | worm | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 6 | 6 | 10 | Invertebrate | clam, crab, crayfish, lobster, octopus, scorpi... | 5 |
| 100 | wren | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | ... | 2 | 1 | 0 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
101 rows × 23 columns
Now we can try to get the “pred” and “class_type” columns to match as best we can. The best way I could come up with to do this is to use value_counts() for both columns, and match them up this way.
df["pred"].value_counts()
0 31
1 20
2 19
3 12
6 8
4 7
5 4
Name: pred, dtype: int64
df["class_type"].value_counts()
0 41
1 20
3 13
6 10
5 8
2 5
4 4
Name: class_type, dtype: int64
I will be matching these values up by creating a dictionary class_dict, matching the numbers in descending order.
class_dict = {0:0, 1:1, 2:3, 3:6, 6:5, 4:2, 5:4}
class_dict
{0: 0, 1: 1, 2: 3, 3: 6, 6: 5, 4: 2, 5: 4}
Testing out some of these values, we can see that the rows corresponding to cluster 1 appear to be birds.
df.loc[(df["pred"] == 1),:]
| animal_name | hair | feathers | eggs | milk | airborne | aquatic | predator | toothed | backbone | ... | legs | tail | domestic | catsize | class_type | Class_Number | Number_Of_Animal_Species_In_Class | Class_Type | Animal_Names | pred | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | chicken | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | ... | 2 | 1 | 1 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
| 16 | crow | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | ... | 2 | 1 | 0 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
| 20 | dove | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | ... | 2 | 1 | 1 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
| 21 | duck | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | ... | 2 | 1 | 0 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
| 23 | flamingo | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | ... | 2 | 1 | 0 | 1 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
| 33 | gull | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | ... | 2 | 1 | 0 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
| 37 | hawk | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | ... | 2 | 1 | 0 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
| 41 | kiwi | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | ... | 2 | 1 | 0 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
| 43 | lark | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | ... | 2 | 1 | 0 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
| 56 | ostrich | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | ... | 2 | 1 | 0 | 1 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
| 57 | parakeet | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | ... | 2 | 1 | 1 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
| 58 | penguin | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | ... | 2 | 1 | 0 | 1 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
| 59 | pheasant | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | ... | 2 | 1 | 0 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
| 71 | rhea | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | ... | 2 | 1 | 0 | 1 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
| 78 | skimmer | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | ... | 2 | 1 | 0 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
| 79 | skua | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | ... | 2 | 1 | 0 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
| 83 | sparrow | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | ... | 2 | 1 | 0 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
| 87 | swan | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | ... | 2 | 1 | 0 | 1 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
| 95 | vulture | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | ... | 2 | 1 | 0 | 1 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
| 100 | wren | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | ... | 2 | 1 | 0 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
20 rows × 23 columns
Similarly, the rows corresponding to cluster 2 appear to be fish.
df.loc[(df["pred"] == 2),:]
| animal_name | hair | feathers | eggs | milk | airborne | aquatic | predator | toothed | backbone | ... | legs | tail | domestic | catsize | class_type | Class_Number | Number_Of_Animal_Species_In_Class | Class_Type | Animal_Names | pred | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | bass | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 3 | 3 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... | 2 |
| 7 | carp | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | ... | 0 | 1 | 1 | 0 | 3 | 3 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... | 2 |
| 8 | catfish | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 3 | 3 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... | 2 |
| 12 | chub | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 3 | 3 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... | 2 |
| 18 | dogfish | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 1 | 3 | 3 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... | 2 |
| 19 | dolphin | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 2 |
| 34 | haddock | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 3 | 3 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... | 2 |
| 38 | herring | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 3 | 3 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... | 2 |
| 60 | pike | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 1 | 3 | 3 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... | 2 |
| 61 | piranha | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 3 | 3 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... | 2 |
| 62 | pitviper | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 2 | 2 | 5 | Reptile | pitviper, seasnake, slowworm, tortoise, tuatara | 2 |
| 66 | porpoise | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 2 |
| 73 | seahorse | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 3 | 3 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... | 2 |
| 74 | seal | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 0 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 2 |
| 76 | seasnake | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 2 | 2 | 5 | Reptile | pitviper, seasnake, slowworm, tortoise, tuatara | 2 |
| 80 | slowworm | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 2 | 2 | 5 | Reptile | pitviper, seasnake, slowworm, tortoise, tuatara | 2 |
| 82 | sole | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 3 | 3 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... | 2 |
| 86 | stingray | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 1 | 3 | 3 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... | 2 |
| 92 | tuna | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 1 | 3 | 3 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... | 2 |
19 rows × 23 columns
Now we can use .map to apply class_dict to the entire row of df["pred"].
df["pred"] = df["pred"].map(class_dict)
df
| animal_name | hair | feathers | eggs | milk | airborne | aquatic | predator | toothed | backbone | ... | legs | tail | domestic | catsize | class_type | Class_Number | Number_Of_Animal_Species_In_Class | Class_Type | Animal_Names | pred | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | aardvark | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 4 | 0 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 |
| 1 | antelope | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | ... | 4 | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 |
| 2 | bass | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 3 | 3 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... | 3 |
| 3 | bear | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 4 | 0 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 |
| 4 | boar | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 4 | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 96 | wallaby | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | ... | 2 | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 2 |
| 97 | wasp | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | ... | 6 | 0 | 0 | 0 | 5 | 5 | 8 | Bug | flea, gnat, honeybee, housefly, ladybird, moth... | 6 |
| 98 | wolf | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 4 | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 |
| 99 | worm | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 6 | 6 | 10 | Invertebrate | clam, crab, crayfish, lobster, octopus, scorpi... | 4 |
| 100 | wren | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | ... | 2 | 1 | 0 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
101 rows × 23 columns
Now that the cluster numbers are somewhat matched up with the class types, we can score these “predictions”. Here we can see that 68/101 clusters match with the class types, which is not a great prediction. We can make graphs using Altair anyway to visualize.
(df["pred"] == df["class_type"]).sum()
68
import altair as alt
I encountered an issue below with the graph. The class types are on the y-axis, with “legs” on the x-axis since it is the characteristic with the largest number of distinct values. The problem with this is that although the points (0,6) and (8,6) are of the same class type, (0,6) is closer in distance to (0,3) than it is to (8,6), and so it gets clustered with (0,3).
alt.Chart(df).mark_circle().encode(
x="legs",
y="class_type",
color="pred:N",
tooltip=["animal_name", "class_type", "pred"]
)
len(numcols)
16
numcols[0]
'hair'
numcols[1]
'feathers'
This is a similar graph using “hair” and “feathers”. Here I encountered my second issue, being that there are only three distinct combinations of these two characteristics - (1,0), (0,0), and (0,1). This means that although there are 101 points, they are all stacked on top of each other at these three points.
alt.Chart(df).mark_circle().encode(
x=numcols[0],
y=numcols[1],
color="pred:N",
tooltip=["animal_name", "class_type", "pred"]
)
Below I attempted to make a list of Altair charts, using “class_type” as the y-axis for all of them and all 16 characteristic columns of df for the x-axis. Using tooltip= allows for information about each point to be displayed when the mouse is hovered over it.
chart_list=[]
for c in range(16):
chart=alt.Chart(df).mark_circle().encode(
x=numcols[c],
y="class_type",
color="pred:N",
tooltip=["animal_name", "class_type", "pred"]
)
chart_list.append(chart)
chart_list[0]
chart_list[1]
The last problem I encountered with these graphs is that when I run chart_list, the charts do not show up.
chart_list
[alt.Chart(...),
alt.Chart(...),
alt.Chart(...),
alt.Chart(...),
alt.Chart(...),
alt.Chart(...),
alt.Chart(...),
alt.Chart(...),
alt.Chart(...),
alt.Chart(...),
alt.Chart(...),
alt.Chart(...),
alt.Chart(...),
alt.Chart(...),
alt.Chart(...),
alt.Chart(...)]
Logistic Regression¶
For the logistic regression section of this project, I will use numcols as the inputs and try to classify Class_Type.
df
| animal_name | hair | feathers | eggs | milk | airborne | aquatic | predator | toothed | backbone | ... | legs | tail | domestic | catsize | class_type | Class_Number | Number_Of_Animal_Species_In_Class | Class_Type | Animal_Names | pred | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | aardvark | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 4 | 0 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 |
| 1 | antelope | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | ... | 4 | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 |
| 2 | bass | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 3 | 3 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... | 3 |
| 3 | bear | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 4 | 0 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 |
| 4 | boar | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 4 | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 96 | wallaby | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | ... | 2 | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 2 |
| 97 | wasp | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | ... | 6 | 0 | 0 | 0 | 5 | 5 | 8 | Bug | flea, gnat, honeybee, housefly, ladybird, moth... | 6 |
| 98 | wolf | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 4 | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 |
| 99 | worm | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 6 | 6 | 10 | Invertebrate | clam, crab, crayfish, lobster, octopus, scorpi... | 4 |
| 100 | wren | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | ... | 2 | 1 | 0 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 |
101 rows × 23 columns
#import
from sklearn.linear_model import LogisticRegression
# instantiate
clf = LogisticRegression()
# fit
clf.fit(df[numcols], df["Class_Type"])
/shared-libs/python3.7/py/lib/python3.7/site-packages/sklearn/linear_model/_logistic.py:818: ConvergenceWarning: lbfgs failed to converge (status=1):
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT.
Increase the number of iterations (max_iter) or scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
Please also refer to the documentation for alternative solver options:
https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression
extra_warning_msg=_LOGISTIC_SOLVER_CONVERGENCE_MSG,
LogisticRegression()
# predict
df["class_pred"] = clf.predict(df[numcols])
df
| animal_name | hair | feathers | eggs | milk | airborne | aquatic | predator | toothed | backbone | ... | tail | domestic | catsize | class_type | Class_Number | Number_Of_Animal_Species_In_Class | Class_Type | Animal_Names | pred | class_pred | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | aardvark | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 0 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 | Mammal |
| 1 | antelope | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | ... | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 | Mammal |
| 2 | bass | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | ... | 1 | 0 | 0 | 3 | 3 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... | 3 | Fish |
| 3 | bear | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 0 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 | Mammal |
| 4 | boar | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 | Mammal |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 96 | wallaby | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | ... | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 2 | Mammal |
| 97 | wasp | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 5 | 5 | 8 | Bug | flea, gnat, honeybee, housefly, ladybird, moth... | 6 | Bug |
| 98 | wolf | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 1 | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 | Mammal |
| 99 | worm | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 6 | 6 | 10 | Invertebrate | clam, crab, crayfish, lobster, octopus, scorpi... | 4 | Invertebrate |
| 100 | wren | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | ... | 1 | 0 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 | Bird |
101 rows × 24 columns
(df["Class_Type"] == df["class_pred"]).sum()
100
This is an example of overfitting the data - 100/101 rows in the dataframe were predicted correctly. Below I wanted to see which one row was predicted incorrectly. The only animal that was predicted incorrectly was “tortoise,” which was predicted as a bird instead of a reptile. My guess is because it lays eggs, which could have confused the computer.
df.loc[(df["Class_Type"] != df["class_pred"]),:]
| animal_name | hair | feathers | eggs | milk | airborne | aquatic | predator | toothed | backbone | ... | tail | domestic | catsize | class_type | Class_Number | Number_Of_Animal_Species_In_Class | Class_Type | Animal_Names | pred | class_pred | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 90 | tortoise | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | ... | 1 | 0 | 1 | 2 | 2 | 5 | Reptile | pitviper, seasnake, slowworm, tortoise, tuatara | 5 | Bird |
1 rows × 24 columns
One thing we can do to combat the overfitting is to split the data into a training set and a testing set. I am using train_size=0.8.
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
df[numcols],
df["Class_Type"],
train_size=0.8
)
X_train
| hair | feathers | eggs | milk | airborne | aquatic | predator | toothed | backbone | breathes | venomous | fins | legs | tail | domestic | catsize | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 48 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 4 | 1 | 0 | 1 |
| 73 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
| 33 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 0 |
| 7 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 |
| 69 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 4 | 1 | 0 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 57 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | 1 | 0 |
| 87 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 1 |
| 62 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
| 66 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| 74 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 |
80 rows × 16 columns
y_train
48 Mammal
73 Fish
33 Bird
7 Fish
69 Mammal
...
57 Bird
87 Bird
62 Reptile
66 Mammal
74 Mammal
Name: Class_Type, Length: 80, dtype: object
# instantiate
clf2 = LogisticRegression()
# fit
clf2.fit(X_train, y_train)
/shared-libs/python3.7/py/lib/python3.7/site-packages/sklearn/linear_model/_logistic.py:818: ConvergenceWarning: lbfgs failed to converge (status=1):
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT.
Increase the number of iterations (max_iter) or scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
Please also refer to the documentation for alternative solver options:
https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression
extra_warning_msg=_LOGISTIC_SOLVER_CONVERGENCE_MSG,
LogisticRegression()
Although it isn’t common practice, we can see that if we predict again on the training set, overfitting is evident once again.
# predict
(clf2.predict(X_train) == y_train).sum()/len(y_train)
1.0
We proceed by predicting on the unseen testing data.
clf2.predict(X_test)
array(['Mammal', 'Mammal', 'Bird', 'Bug', 'Fish', 'Mammal', 'Mammal',
'Bug', 'Mammal', 'Mammal', 'Bird', 'Mammal', 'Mammal', 'Bug',
'Bird', 'Invertebrate', 'Mammal', 'Fish', 'Mammal', 'Mammal',
'Mammal'], dtype=object)
I will now calculate the score both manually and by using clf2.score. This is a pretty good score; 95.2% of the rows were predicted correctly. While there may be some overfitting still, it is much less evident fitting on the training set and predicting on the testing data.
(clf2.predict(X_test) == y_test).sum()/len(y_test)
0.9523809523809523
# same as:
clf2.score(X_test, y_test)
0.9523809523809523
Decision Tree¶
The last section from the course material is decision trees. My goal here is to see which characteristics are the most “important” in determining classification. We will try first without using test or train sets for comparison.
# import
from sklearn.tree import DecisionTreeClassifier
# instantiate
clf_tree = DecisionTreeClassifier(max_depth=4, max_leaf_nodes=10)
# fit
clf_tree.fit(df[numcols], df["Class_Type"])
DecisionTreeClassifier(max_depth=4, max_leaf_nodes=10)
from sklearn import tree
import matplotlib.pyplot as plt
# plot
fig = plt.figure(figsize=(200,100))
_ = tree.plot_tree(clf_tree,
feature_names=clf_tree.feature_names_in_,
class_names=clf_tree.classes_,
filled=True)
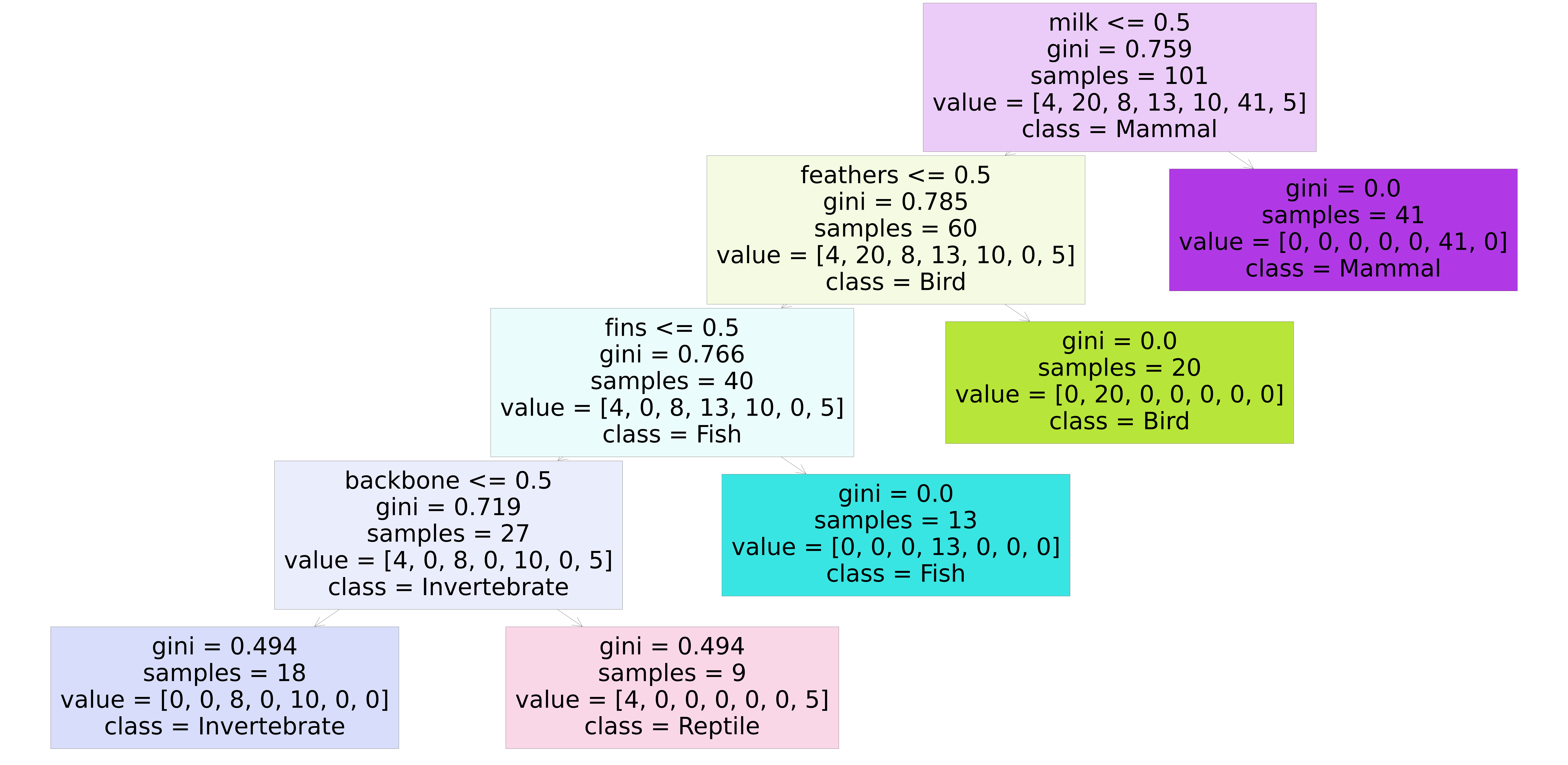
This diagram shows us the most important characteristics for each class type. For mammals it’s milk, for birds it’s feathers, etc. It seems like there may be a bit of overfitting since “mammal”, “bird”, and “fish” all have 100% probabilities, however the rest do not.
Now we can do the same thing with X_train and y_train to compare the two.
# instantiate
clf_tree2 = DecisionTreeClassifier(max_depth=4, max_leaf_nodes=12)
# fit
clf_tree2.fit(X_train, y_train)
DecisionTreeClassifier(max_depth=4, max_leaf_nodes=12)
# plot
fig = plt.figure(figsize=(200,100))
_ = tree.plot_tree(clf_tree2,
feature_names=clf_tree2.feature_names_in_,
class_names=clf_tree2.classes_,
filled=True)
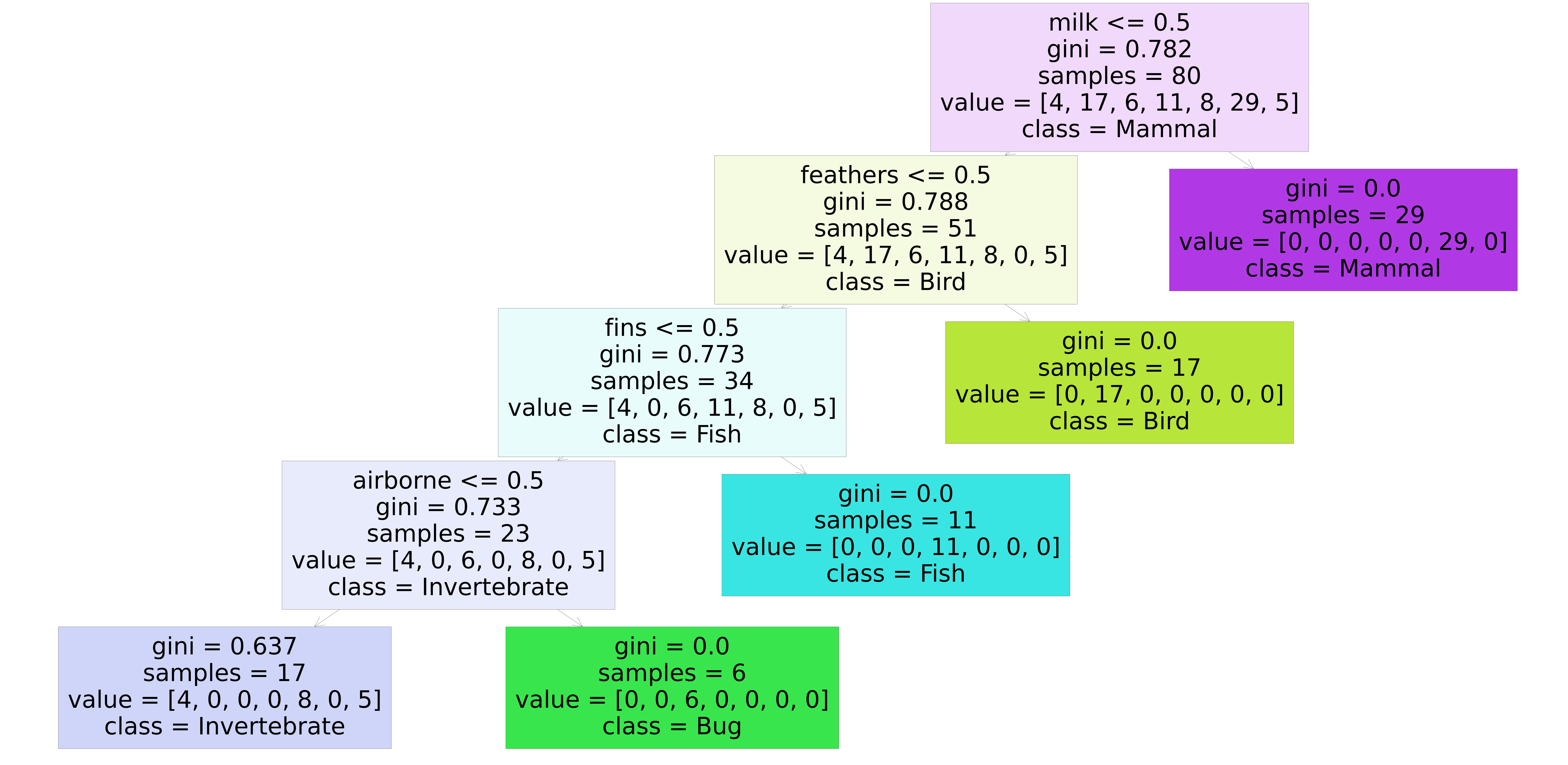
The two plots are identical except for the bottom two leve;s; in the top one it is “backbone,” while in the second one it is “airborne”. Now we can use score to compare the two.
clf_tree.score(df[numcols], df["Class_Type"])
0.8811881188118812
clf_tree2.score(X_train, y_train)
0.8875
We can see that the decision tree classifier does slightly better when testing and training sets are used, but it isn’t too big of a difference. Overall, the score for the decision tree was lower than that of logistic regression.
K-Nearest Neighbors¶
# import
from sklearn.neighbors import KNeighborsClassifier
# instantiate
clf_nb = KNeighborsClassifier(n_neighbors=10)
Since I specified n_neighbors=10 during the instantiating step, this is going to find the 10 nearest data points for our new data point based on distance, similar to K-Means clustering. However, for K-Nearest Neighbors we will be trying to predict “Class_Type” once again.
clf_nb.fit(df[numcols], df["Class_Type"])
KNeighborsClassifier(n_neighbors=10)
df["pred_nb"] = clf_nb.predict(df[numcols])
df
| animal_name | hair | feathers | eggs | milk | airborne | aquatic | predator | toothed | backbone | ... | domestic | catsize | class_type | Class_Number | Number_Of_Animal_Species_In_Class | Class_Type | Animal_Names | pred | class_pred | pred_nb | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | aardvark | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 | Mammal | Mammal |
| 1 | antelope | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 | Mammal | Mammal |
| 2 | bass | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 0 | 3 | 3 | 13 | Fish | bass, carp, catfish, chub, dogfish, haddock, h... | 3 | Fish | Fish |
| 3 | bear | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 | Mammal | Mammal |
| 4 | boar | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 | Mammal | Mammal |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 96 | wallaby | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 2 | Mammal | Mammal |
| 97 | wasp | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 5 | 5 | 8 | Bug | flea, gnat, honeybee, housefly, ladybird, moth... | 6 | Bug | Bug |
| 98 | wolf | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 0 | Mammal | Mammal |
| 99 | worm | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 6 | 6 | 10 | Invertebrate | clam, crab, crayfish, lobster, octopus, scorpi... | 4 | Invertebrate | Fish |
| 100 | wren | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | ... | 0 | 0 | 1 | 1 | 20 | Bird | chicken, crow, dove, duck, flamingo, gull, haw... | 1 | Bird | Bird |
101 rows × 25 columns
After adding a new column to df with the prediction from K-Nearest Neighbors, we can use score to compare this to the other methods.
clf_nb.score(df[numcols], df["Class_Type"])
0.8316831683168316
This score is not the best, but it’s not bad either. Here I am taking a closer look at exactly which columns were difficult for the computer to predict. It seems like the invertebrates were especially challenging to predict using this method.
df.loc[(df["Class_Type"] != df["pred_nb"]), :]
| animal_name | hair | feathers | eggs | milk | airborne | aquatic | predator | toothed | backbone | ... | domestic | catsize | class_type | Class_Number | Number_Of_Animal_Species_In_Class | Class_Type | Animal_Names | pred | class_pred | pred_nb | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13 | clam | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | ... | 0 | 0 | 6 | 6 | 10 | Invertebrate | clam, crab, crayfish, lobster, octopus, scorpi... | 4 | Invertebrate | Fish |
| 14 | crab | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | ... | 0 | 0 | 6 | 6 | 10 | Invertebrate | clam, crab, crayfish, lobster, octopus, scorpi... | 5 | Invertebrate | Amphibian |
| 15 | crayfish | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | ... | 0 | 0 | 6 | 6 | 10 | Invertebrate | clam, crab, crayfish, lobster, octopus, scorpi... | 6 | Invertebrate | Bug |
| 19 | dolphin | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 3 | Mammal | Fish |
| 46 | lobster | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | ... | 0 | 0 | 6 | 6 | 10 | Invertebrate | clam, crab, crayfish, lobster, octopus, scorpi... | 6 | Invertebrate | Bug |
| 53 | octopus | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | ... | 0 | 1 | 6 | 6 | 10 | Invertebrate | clam, crab, crayfish, lobster, octopus, scorpi... | 6 | Invertebrate | Bug |
| 62 | pitviper | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | ... | 0 | 0 | 2 | 2 | 5 | Reptile | pitviper, seasnake, slowworm, tortoise, tuatara | 3 | Reptile | Fish |
| 66 | porpoise | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 3 | Mammal | Fish |
| 72 | scorpion | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | ... | 0 | 0 | 6 | 6 | 10 | Invertebrate | clam, crab, crayfish, lobster, octopus, scorpi... | 6 | Invertebrate | Bug |
| 74 | seal | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 1 | 0 | 0 | 41 | Mammal | aardvark, antelope, bear, boar, buffalo, calf,... | 3 | Mammal | Fish |
| 76 | seasnake | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | ... | 0 | 0 | 2 | 2 | 5 | Reptile | pitviper, seasnake, slowworm, tortoise, tuatara | 3 | Reptile | Fish |
| 77 | seawasp | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | ... | 0 | 0 | 6 | 6 | 10 | Invertebrate | clam, crab, crayfish, lobster, octopus, scorpi... | 4 | Invertebrate | Fish |
| 80 | slowworm | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | ... | 0 | 0 | 2 | 2 | 5 | Reptile | pitviper, seasnake, slowworm, tortoise, tuatara | 3 | Reptile | Fish |
| 81 | slug | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 6 | 6 | 10 | Invertebrate | clam, crab, crayfish, lobster, octopus, scorpi... | 4 | Invertebrate | Fish |
| 90 | tortoise | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | ... | 0 | 1 | 2 | 2 | 5 | Reptile | pitviper, seasnake, slowworm, tortoise, tuatara | 5 | Bird | Mammal |
| 91 | tuatara | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | ... | 0 | 0 | 2 | 2 | 5 | Reptile | pitviper, seasnake, slowworm, tortoise, tuatara | 5 | Reptile | Amphibian |
| 99 | worm | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 6 | 6 | 10 | Invertebrate | clam, crab, crayfish, lobster, octopus, scorpi... | 4 | Invertebrate | Fish |
17 rows × 25 columns
Now we can add in our X_train, y_train to fit and X_test, y_test to score.
clf_nb2 = KNeighborsClassifier(n_neighbors=10)
clf_nb2.fit(X_train, y_train)
KNeighborsClassifier(n_neighbors=10)
clf_nb2.score(X_train, y_train)
0.8
clf_nb2.score(X_test, y_test)
0.9047619047619048
I find it a bit strange that the testing set hada much better score than the train set, and both scores are still lower than that of both logistic regression and the decision tree classifier.
Summary¶
In summary, I used K-Means clustering, logistic regression, decision tree classifiers, and K-Nearest Neighbors to attempt to classify zoo animals. The results show that logistic regression was best (with possibility of overfitting), followed by decision tree classifiers, and then K-Nearest Neighbors. K-Means clustering was not the best suited for this project, but was still a good tool to visualize.
References¶
What is the source of your dataset(s)?
Were any portions of the code or ideas taken from another source? List those sources here and say how they were used.
I used this article from analyticsvidhya.com to come up with the K-Nearest Neighbors portion of my project.
