Week 6 Videos
Contents
Week 6 Videos#
Artificial data for linear regression#
import pandas as pd
import altair as alt
from sklearn.datasets import make_regression
help(make_regression)
Help on function make_regression in module sklearn.datasets._samples_generator:
make_regression(n_samples=100, n_features=100, *, n_informative=10, n_targets=1, bias=0.0, effective_rank=None, tail_strength=0.5, noise=0.0, shuffle=True, coef=False, random_state=None)
Generate a random regression problem.
The input set can either be well conditioned (by default) or have a low
rank-fat tail singular profile. See :func:`make_low_rank_matrix` for
more details.
The output is generated by applying a (potentially biased) random linear
regression model with `n_informative` nonzero regressors to the previously
generated input and some gaussian centered noise with some adjustable
scale.
Read more in the :ref:`User Guide <sample_generators>`.
Parameters
----------
n_samples : int, default=100
The number of samples.
n_features : int, default=100
The number of features.
n_informative : int, default=10
The number of informative features, i.e., the number of features used
to build the linear model used to generate the output.
n_targets : int, default=1
The number of regression targets, i.e., the dimension of the y output
vector associated with a sample. By default, the output is a scalar.
bias : float, default=0.0
The bias term in the underlying linear model.
effective_rank : int, default=None
If not None:
The approximate number of singular vectors required to explain most
of the input data by linear combinations. Using this kind of
singular spectrum in the input allows the generator to reproduce
the correlations often observed in practice.
If None:
The input set is well conditioned, centered and gaussian with
unit variance.
tail_strength : float, default=0.5
The relative importance of the fat noisy tail of the singular values
profile if `effective_rank` is not None. When a float, it should be
between 0 and 1.
noise : float, default=0.0
The standard deviation of the gaussian noise applied to the output.
shuffle : bool, default=True
Shuffle the samples and the features.
coef : bool, default=False
If True, the coefficients of the underlying linear model are returned.
random_state : int, RandomState instance or None, default=None
Determines random number generation for dataset creation. Pass an int
for reproducible output across multiple function calls.
See :term:`Glossary <random_state>`.
Returns
-------
X : ndarray of shape (n_samples, n_features)
The input samples.
y : ndarray of shape (n_samples,) or (n_samples, n_targets)
The output values.
coef : ndarray of shape (n_features,) or (n_features, n_targets)
The coefficient of the underlying linear model. It is returned only if
coef is True.
make_regression(n_samples=4, n_features=2)
(array([[ 1.44089066, -2.55372173],
[ 0.65938893, 0.91995171],
[-1.82165048, -0.80337151],
[ 0.03900098, -0.45748574]]),
array([-88.39997798, 32.6088018 , -29.28375752, -15.97838099]))
X, y = make_regression(n_samples=50, n_features=1)
X
array([[-0.15025868],
[-0.42176296],
[-0.29864099],
[ 0.32309439],
[ 0.90403753],
[-0.24620504],
[ 0.44303931],
[-0.45439768],
[ 0.93924799],
[ 1.20805558],
[ 0.38826733],
[-0.67992318],
[-0.819682 ],
[-0.18220005],
[-0.1647296 ],
[-0.27437199],
[ 1.52415097],
[ 0.80834783],
[ 0.54360419],
[ 1.33736049],
[-2.22028198],
[-0.53716202],
[-3.33133071],
[ 0.02567355],
[-0.90405335],
[-1.26884617],
[-3.03271921],
[-0.87246159],
[-0.86043787],
[ 0.83300092],
[-1.52452591],
[-0.5437913 ],
[ 2.17092195],
[-0.50819959],
[-0.84526105],
[ 0.03153603],
[-0.9748441 ],
[-0.45361931],
[-0.43091817],
[-0.57319906],
[-0.63619132],
[ 0.75722195],
[ 0.71109085],
[-1.38667905],
[-0.04297198],
[ 0.42028346],
[-0.2517879 ],
[ 2.67056034],
[-0.17628291],
[-1.95496794]])
X.shape
(50, 1)
y
array([ -13.43582581, -37.71318709, -26.7038709 , 28.89044381,
80.83720026, -22.01515484, 39.61567569, -40.63131744,
83.98564772, 108.02187692, 34.71807587, -60.79735032,
-73.29430019, -16.29195804, -14.72978653, -24.53378629,
136.28648463, 72.28082116, 48.60798302, 119.58405937,
-198.53310623, -48.03193734, -297.88082665, 2.29567689,
-80.8386151 , -113.45764779, -271.17959352, -78.01374335,
-76.93860658, 74.48525081, -136.32001068, -48.62471396,
194.11943247, -45.44217495, -75.58152675, 2.81988733,
-87.16857999, -40.56171776, -38.53182755, -51.25429569,
-56.88693572, 67.70924927, 63.58429958, -123.99402449,
-3.84246716, 37.58089326, -22.51436272, 238.79608281,
-15.76286018, -174.8092634 ])
y.shape
(50,)
pd.DataFrame({"col0": X, "col1": y})
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In [11], line 1
----> 1 pd.DataFrame({"col0": X, "col1": y})
File /shared-libs/python3.9/py/lib/python3.9/site-packages/pandas/core/frame.py:529, in DataFrame.__init__(self, data, index, columns, dtype, copy)
524 mgr = self._init_mgr(
525 data, axes={"index": index, "columns": columns}, dtype=dtype, copy=copy
526 )
528 elif isinstance(data, dict):
--> 529 mgr = init_dict(data, index, columns, dtype=dtype)
530 elif isinstance(data, ma.MaskedArray):
531 import numpy.ma.mrecords as mrecords
File /shared-libs/python3.9/py/lib/python3.9/site-packages/pandas/core/internals/construction.py:287, in init_dict(data, index, columns, dtype)
281 arrays = [
282 arr if not isinstance(arr, ABCIndexClass) else arr._data for arr in arrays
283 ]
284 arrays = [
285 arr if not is_datetime64tz_dtype(arr) else arr.copy() for arr in arrays
286 ]
--> 287 return arrays_to_mgr(arrays, data_names, index, columns, dtype=dtype)
File /shared-libs/python3.9/py/lib/python3.9/site-packages/pandas/core/internals/construction.py:85, in arrays_to_mgr(arrays, arr_names, index, columns, dtype, verify_integrity)
82 index = ensure_index(index)
84 # don't force copy because getting jammed in an ndarray anyway
---> 85 arrays = _homogenize(arrays, index, dtype)
87 columns = ensure_index(columns)
88 else:
File /shared-libs/python3.9/py/lib/python3.9/site-packages/pandas/core/internals/construction.py:355, in _homogenize(data, index, dtype)
353 val = dict(val)
354 val = lib.fast_multiget(val, oindex._values, default=np.nan)
--> 355 val = sanitize_array(
356 val, index, dtype=dtype, copy=False, raise_cast_failure=False
357 )
359 homogenized.append(val)
361 return homogenized
File /shared-libs/python3.9/py/lib/python3.9/site-packages/pandas/core/construction.py:529, in sanitize_array(data, index, dtype, copy, raise_cast_failure)
527 elif subarr.ndim > 1:
528 if isinstance(data, np.ndarray):
--> 529 raise ValueError("Data must be 1-dimensional")
530 else:
531 subarr = com.asarray_tuplesafe(data, dtype=dtype)
ValueError: Data must be 1-dimensional
df = pd.DataFrame({"col0": X.reshape(-1), "col1": y})
alt.Chart(df).mark_circle().encode(
x="col0",
y="col1"
)
Using noise and bias in make_regression#
def make_chart(X,y):
df = pd.DataFrame({"col0": X.reshape(-1), "col1": y})
chart = alt.Chart(df).mark_circle().encode(
x="col0",
y="col1"
)
return chart
X, y = make_regression(n_samples=50, n_features=1, noise=10, bias=-14.5)
make_chart(X,y)
X, y = make_regression(n_samples=50, n_features=1, noise=10, bias=-14.5)
make_chart(X,y)
X, y = make_regression(n_samples=50, n_features=1, noise=10, bias=-14.5, random_state=1234)
make_chart(X,y)
X, y = make_regression(n_samples=50, n_features=1, noise=10, bias=-14.5, random_state=1234, coef=True)
make_chart(X,y)
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In [42], line 1
----> 1 X, y = make_regression(n_samples=50, n_features=1, noise=10, bias=-14.5, random_state=1234, coef=True)
2 make_chart(X,y)
ValueError: too many values to unpack (expected 2)
X, y, m = make_regression(n_samples=50, n_features=1, noise=10, bias=-14.5, random_state=1234, coef=True)
make_chart(X,y)
m
array(79.05241331)
Estimating the coefficient and bias using LinearRegression from scikit-learn#
X, y, m = make_regression(n_samples=50, n_features=1, noise=10, bias=-14.5, random_state=1234, coef=True)
make_chart(X,y)
from sklearn.linear_model import LinearRegression
reg = LinearRegression()
type(reg)
sklearn.linear_model._base.LinearRegression
reg.coef_
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In [18], line 1
----> 1 reg.coef_
AttributeError: 'LinearRegression' object has no attribute 'coef_'
reg.intercept_
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In [19], line 1
----> 1 reg.intercept_
AttributeError: 'LinearRegression' object has no attribute 'intercept_'
reg.fit(X,y)
LinearRegression()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
LinearRegression()
reg.coef_
array([80.25327575])
reg.intercept_
-12.675241213173738
Plotting using Matplotlib#
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
ax.plot(X,y)
[<matplotlib.lines.Line2D at 0x7fec7f4bdfd0>]
fig, ax = plt.subplots()
ax.plot(X,y, 'kx')
[<matplotlib.lines.Line2D at 0x7fec7f319280>]
fig, ax = plt.subplots()
ax.scatter(X,y)
<matplotlib.collections.PathCollection at 0x7fecade860a0>
plt.style.available
['Solarize_Light2',
'_classic_test_patch',
'_mpl-gallery',
'_mpl-gallery-nogrid',
'bmh',
'classic',
'dark_background',
'fast',
'fivethirtyeight',
'ggplot',
'grayscale',
'seaborn-v0_8',
'seaborn-v0_8-bright',
'seaborn-v0_8-colorblind',
'seaborn-v0_8-dark',
'seaborn-v0_8-dark-palette',
'seaborn-v0_8-darkgrid',
'seaborn-v0_8-deep',
'seaborn-v0_8-muted',
'seaborn-v0_8-notebook',
'seaborn-v0_8-paper',
'seaborn-v0_8-pastel',
'seaborn-v0_8-poster',
'seaborn-v0_8-talk',
'seaborn-v0_8-ticks',
'seaborn-v0_8-white',
'seaborn-v0_8-whitegrid',
'tableau-colorblind10']
plt.style.use('ggplot')
fig, ax = plt.subplots()
ax.scatter(X,y)
<matplotlib.collections.PathCollection at 0x7fecade66f10>
import numpy as np
Xtrue = np.array([-2,2]).reshape(-1,1)
ytrue = reg.predict(Xtrue)
ytrue
array([-173.18179271, 147.83131029])
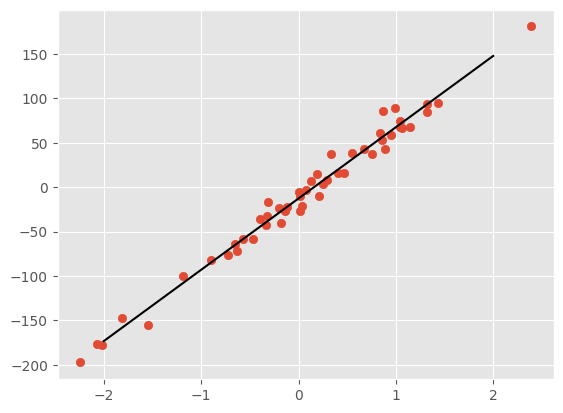
fig, ax = plt.subplots()
ax.scatter(X,y)
ax.plot(Xtrue, ytrue, 'k')
[<matplotlib.lines.Line2D at 0x7fecad723bb0>]
reg.predict(np.array([-2,2]))
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In [37], line 1
----> 1 reg.predict(np.array([-2,2]))
File /shared-libs/python3.9/py/lib/python3.9/site-packages/sklearn/linear_model/_base.py:386, in LinearModel.predict(self, X)
372 def predict(self, X):
373 """
374 Predict using the linear model.
375
(...)
384 Returns predicted values.
385 """
--> 386 return self._decision_function(X)
File /shared-libs/python3.9/py/lib/python3.9/site-packages/sklearn/linear_model/_base.py:369, in LinearModel._decision_function(self, X)
366 def _decision_function(self, X):
367 check_is_fitted(self)
--> 369 X = self._validate_data(X, accept_sparse=["csr", "csc", "coo"], reset=False)
370 return safe_sparse_dot(X, self.coef_.T, dense_output=True) + self.intercept_
File /shared-libs/python3.9/py/lib/python3.9/site-packages/sklearn/base.py:577, in BaseEstimator._validate_data(self, X, y, reset, validate_separately, **check_params)
575 raise ValueError("Validation should be done on X, y or both.")
576 elif not no_val_X and no_val_y:
--> 577 X = check_array(X, input_name="X", **check_params)
578 out = X
579 elif no_val_X and not no_val_y:
File /shared-libs/python3.9/py/lib/python3.9/site-packages/sklearn/utils/validation.py:879, in check_array(array, accept_sparse, accept_large_sparse, dtype, order, copy, force_all_finite, ensure_2d, allow_nd, ensure_min_samples, ensure_min_features, estimator, input_name)
877 # If input is 1D raise error
878 if array.ndim == 1:
--> 879 raise ValueError(
880 "Expected 2D array, got 1D array instead:\narray={}.\n"
881 "Reshape your data either using array.reshape(-1, 1) if "
882 "your data has a single feature or array.reshape(1, -1) "
883 "if it contains a single sample.".format(array)
884 )
886 if dtype_numeric and array.dtype.kind in "USV":
887 raise ValueError(
888 "dtype='numeric' is not compatible with arrays of bytes/strings."
889 "Convert your data to numeric values explicitly instead."
890 )
ValueError: Expected 2D array, got 1D array instead:
array=[-2 2].
Reshape your data either using array.reshape(-1, 1) if your data has a single feature or array.reshape(1, -1) if it contains a single sample.
reg.predict(np.array([-2,2]).reshape(-1,1))
array([-173.18179271, 147.83131029])
